
The surprising importance of gut microbiota in tackling flu infection
Our understanding of defence mechanisms raised against a given pathogen (e.g., influenza virus infection) is mainly focused on innate and adaptive immune cells responses, but other cell types such as stromal cells most certainly constitute an important first line of defence against invading pathogens. In addition to creating a primary physical barrier, stromal cells are armed with intrinsic immune defence mechanisms that cover a broad set of antiviral mechanisms including autophagy, inflammasome response, and the interferon (IFN) system. Cell autonomous immunity is tightly regulated, and needs to be unlocked by inflammatory signals including type-I interferon (IFN-I). Since the IFN-I receptor (composed of IFNAR1 and IFNAR2 subunits) is ubiquitously expressed, IFN-I acts as an alarm signal on both stromal and hematopoietic cells. Following binding to its cognate receptor, IFN-I elicits a potent antiviral state by inducing the expression of hundreds of genes collectively called IFN-stimulated genes (ISGs). Some ISGs carry intrinsic antiviral functions that were shown to dampen influenza virus replication including MX1, IFITM3, and NCOA7 among the most significant (1,2). Therefore, once established, the IFN-induced antiviral state endows the cells with the capacity to limit the replication and spread of several classes of viruses including influenza virus (3,4). Acute IFN-I production is triggered following sensing of invading pathogens by pattern recognition receptors (PRRs) that signal through MyD88, TRIF, MAVS or STING in order to activate the master regulators of IFN-I production, IRF3 and IRF7 transcription factors (5). However, this acute IFN production requires prior cell infection and is induced after a first wave of viral replication, therefore it cannot confer immediate antiviral defences. Moreover, viruses have evolved different strategies to avoid sensing and limit IFN induction. For instance, the NS1 protein from influenza virus acts through multiple anti-IFN strategies (6). Interestingly, in contrast to acute IFN production, pioneer observations reported a steady-state basal IFN production in absence of infection [reviewed in (7)]. This constitutive IFN production has gained much attention over the last decade and has revealed important roles in the regulation of immune cell function and homeostasis as well as for the maintenance of basal antiviral and antitumor immunity (7). Of note, dysregulated or chronic IFN production is associated with several pathological outcomes (8). Thus, IFN production requires a tight regulation in terms of timing and quantity. We can therefore ask, what are the molecular determinants that drive basal IFN production? How is it regulated by the host? By which mechanism does the steady-state IFN signature contribute to set basal antiviral defences? In a recent issue of Cell Reports, Bradley et al. (9) provided substantial answers to these burning questions (Figure 1).
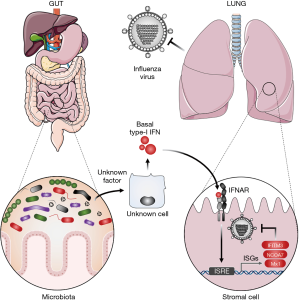
Bradley et al. (9) made the crucial observation that uninfected naïve mice present a significant level of ISG expression (Oasl2, Rsad2) in the lung at steady-state, which is critically decreased in IFNAR1−/− mice or in mice treated with an anti-IFNAR antibody. Taking advantage of a genetically engineered mice model in which IFNAR1 downregulation (otherwise occurring rapidly upon IFN stimulation) is impaired by a single mutation (IFNAR1SA), the authors could significantly increase basal ISG expression in lung cells. Using this elegant model, the authors could draw two important conclusions. First, they provided further evidence that tonic IFN-I production is responsible for basal ISG expression in the lungs of uninfected mice, as previously reported by other groups (10,11). Second, this basal IFN response is tightly regulated through downregulation of cell surface IFNAR expression (12).
Immunological relevance of this basal IFN signature came from the demonstration that IFNAR1SA mice show better resistance to influenza virus infection with a significant decrease in viral gene expression and infection-induced weight loss. It is remarkable that tuning basal IFN signature can help to boost the steady-state antiviral defences in vivo.
Interestingly, among lung resident cells, basal IFN-signatures can be found not only in CD45-positive, immune cells [as previously reported for macrophage populations (10)] but also in CD45-negative, non-immune cells, in an even more pronounced way. This led the authors to question a possible role for stromal cells in the control of influenza virus infection. Accordingly, key hematopoietic chimera experiments using either wild type (WT) (C57BL/6) mice reconstituted with IFNAR1SA hematopoietic cells, or conversely IFNAR1SA reconstituted with WT hematopoietic cells, highlighted a fundamental role for radio-resistant stromal cells in the early dampening of influenza virus infection. More precisely, Bradley et al. demonstrated that basal IFN-signature activates two sequential immune protections against influenza virus. An early first line of defence that is significantly supported by stromal cells (significant at day 2 p.i.). Later, a second protection is conferred by immune cells (significant at day 8 p.i.). This result highlighted a dual role for tonic IFN signature to sustain both stromal and immune cell defences that cooperate to confer optimal protection against influenza virus infection.
Such observations raised critical questions about the host determinants that are driving basal IFN-I production. Because plasmacytoid dendritic cells (pDC) are the major source of IFN-I, the authors tested the implication of these cells in basal IFN-I production. Surprisingly, pDC depletion by an anti-PDCA1 antibody-mediated approach did not alter basal IFN-signature in the whole lung, suggesting that another source (still unidentified) is crucial to drive basal IFN-response. In order to help identify this source, it would be interesting to determine which IFN-I subtype is responsible for tonic IFN-response in the lung. Indeed, the different IFN-I subtypes (i.e., -α, -β, -ε, -κ, -ω) may be produced by different cells and subjected to different regulatory mechanisms (13). For instance, IFN-ε is constitutively expressed at mucosal surfaces and was shown to be regulated by hormones rather that pathogen sensors (13).
Previous study by Abt et al. identified an essential role for microbiota in driving basal ISG signatures in macrophages (10). To investigate a potential role for microbiota at inducing basal ISG expression in stromal cells, Bradley et al. compared basal ISG expression in lungs of mice that were fed with a cocktail of antibiotics (ampicillin, vancomycin, metronidazole, gentamicin) or plain-water for 3–4 weeks. Remarkably, the authors observed a drastic decrease of ISG expression in the CD45-negative compartment of antibiotic-treated mice. This observation thus broadens the impact of microbiota in driving basal IFN-I production in the stromal cell compartment, in addition to the previously described effects on immune cells and particularly on macrophages (10).
Microbiota is an integral part of animals, which is composed of host-residing bacteria, viruses, fungi, protozoa and archaea that colonize all mucosal surfaces including respiratory, urogenital, and gastro-intestinal tracts in which the microbial population is the most abundant. Because orally taken antibiotics are known to systemically impact microbiota composition, the authors questioned the origin of the microbiota responsible for basal IFN-I production. Importantly, they show that faecal transplantation was able to rescue basal ISG signature in stromal cells of antibiotic treated-mice, arguing for bacterial signals coming from the gut. It is hence quite fascinating that local gut microbiota can influence immunity at distal sites including the lung. This important cross-talk was previously described as the gut-lung axis (14).
Microbiota produce a wealth of microbe-associated molecular pattern (MAMP), toxins and metabolites including short chain fatty acids (SCFA), a product of dietary fiber fermentation, that was shown to regulate immune cell metabolism, activation status and homeostasis (14). In order to identify microbial determinants that drive tonic IFN-signature, Steed et al. (11) screened a library of 84 microbial metabolites for their ability to trigger or amplify type-I IFN response and found that desaminotyrosine (DAT), a degradation product of flavonoid, had a significant effect. Mechanistically, the authors demonstrated that DAT enhanced IFN responses through amplifying STAT1 signalling, but not through induction of IFN-I production. Interestingly, authors hypothesized that the endogenous virus component of the microbiota could be responsible for initial IFN-I induction. Thus, it is tempting to speculate that one of these previously described microbiota derived components might be responsible for basal ISG expression in the stromal cells reported by Bradley et al. However, unravelling the precise mechanism will warrant further work.
Among the ISGs that are able to restrict influenza infection, Mx1 codes a dynamin-like large GTPase and appears to be one of the most potent in mice (15). However, most inbred mice, including C57BL/6 used in this study, have lost Mx1 gene expression. Thus, a step towards the translational importance of their findings came from the use of a genetically engineered mice model in which Mx1 gene, close homologous to human MxA, was introduced. Importantly, Mx1 transgenic mice demonstrated sustained microbiota-driven tonic IFN levels that conferred basal protection against influenza, as shown by the negative impact of antibiotics on the outcome of infection.
Prior notable reports had paved the way for this fascinating study. Ichinohe et al. (16) demonstrated that antibiotic-treated mice failed to develop efficient innate and adaptive immune responses against influenza virus. This defect was not attributed to a general immunodeficiency, nor was it specific to lung infections. Indeed, antibiotic-treated mice mounted comparable Ig and T-cell responses to mice with a native microbiota in response to footpad immunization with ovalbumin or intranasal infection with either herpes simplex virus type 2 (HSV-2) or Legionella pneumophila. Interestingly, this defect was associated with impaired inflammasome response (decreased level of bronchoalveolar lavage IL-18 and IL-1β) in the lung and could be restored following local (intranasal) or distal (rectal) administration of Toll-like receptor (TLR) ligands. These findings highlighted an important impact for the microbiota to influence immunity at distal sites through modulation of the inflammasome response. In continuity with this report, Abt et al. (10) observed a similar defect in innate and adaptive immune responses to influenza infection or systemic lymphocytic choriomeningitis virus (LCMV) infection of germ-free or antibiotic-treated mice. To identify a potential mechanism through which signals derived from commensal bacteria regulate macrophage responses, Abt et al. performed a genome-wide transcriptional profiling and found diminished expression of IFN-related genes in macrophages isolated from antibiotic-treated mice. However, IFN signature in non-immune cells was not investigated. In line with these previous reports, Bradley et al. (9) unravelled that microbiota is driving steady-state IFN-signature in non-immune, stromal cells and thus broadens the complex interplay between gut microbiota and lung immune responses to influenza virus.
Inflammasome response is essential to confer protective immunity against influenza infection (17). Specifically, influenza virus has been shown to activate the NLRP3 inflammasome in macrophages, but not in lung epithelial cells, through the ion channel-forming M2 protein (18). Although NLRP3 sensor is not expressed in lung epithelial cells, recent data from Lee et al. (19) demonstrated that IFN-I-induced MxA (the human homologous of mice Mx1) can substitute NLRP3 in order to restore optimal inflammasome responses of lung epithelial cells and host resistance to influenza virus. Since IFN is known to unlock the inflammasome response through induction of guanylate-binding proteins (GBPs) (20), inflammatory caspases 4/5/11 (21) and MxA (19), it might be tempting to speculate that both IFN and inflammasome responses are playing contributing roles in microbiota-driven anti-influenza immune defence of lung stromal cells. This will nevertheless require further investigation.
This study raised a number of interesting questions. What are the precise commensal species, or components responsible for boosting basal immune defences? Answering this question is challenging since most of gut-residing bacterial species remain largely unstudied due to lack of suitable culture conditions. Substantial progress to potentially overcome this obstacle was recently published (22) where authors identified γ-aminobutyric acid (GABA) as an important bacterial growth factor. Additionally, the role of non-bacterial species of microbiota including virus, fungi, and parasites remained largely unexplored. When considering the impact of gut microbiota in tuning basal anti-viral responses, it will be important to determine whether specific diet or probiotic therapy can be explored to boost immune protection during the flu season. Conversely, precaution must be put forward against systematic taking of antibiotics, which can alter rather than improve the state of health. Interestingly, recent studies demonstrated that antibiotic-usage impairs immunogenicity and response to influenza vaccination in human (23). In conclusion, manipulation of microbiota in order to boost basal stromal cell defences and sustain antiviral immunity may pave the way to future therapeutic strategies.
Acknowledgments
The authors wish to thank Joe McKellar for his useful comments on the manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Executive Editor-in-Chief Dr. Hualin Sun (Jiangsu Key Laboratory of Neuroregeneration, Nantong University, Nantong, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2020.03.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol 2014;6:40-6. [Crossref] [PubMed]
- Doyle T, Moncorge O, Bonaventure B, et al. The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry. Nat Microbiol 2018;3:1369-76. [Crossref] [PubMed]
- Richman DD, Murphy BR, Baron S, et al. Three strains of influenza A virus (H3N2): interferon sensitivity in vitro and interferon production in volunteers. J Clin Microbiol 1976;3:223-6. [PubMed]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 1957;147:258-67. [Crossref] [PubMed]
- Trinchieri G. Type I interferon: friend or foe? J Exp Med 2010;207:2053-63. [Crossref] [PubMed]
- Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 2007;81:7011-21. [Crossref] [PubMed]
- Gough DJ, Messina NL, Clarke CJ, et al. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 2012;36:166-74. [Crossref] [PubMed]
- Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci 2011;1238:91-8. [Crossref] [PubMed]
- Bradley KC, Finsterbusch K, Schnepf D, et al. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep 2019;28:245-256.e4. [Crossref] [PubMed]
- Abt MC, Osborne LC, Monticelli LA, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012;37:158-70. [Crossref] [PubMed]
- Steed AL, Christophi GP, Kaiko GE, et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 2017;357:498-502. [Crossref] [PubMed]
- Bhattacharya S, Katlinski KV, Reichert M, et al. Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol Med 2014;6:384-97. [Crossref] [PubMed]
- Li SF, Gong MJ, Zhao FR, et al. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cell Physiol Biochem 2018;51:2377-96. [Crossref] [PubMed]
- Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol 2017;15:55-63. [Crossref] [PubMed]
- Haller O, Stertz S, Kochs G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect 2007;9:1636-43. [Crossref] [PubMed]
- Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 2011;108:5354-9. [Crossref] [PubMed]
- Ichinohe T, Lee HK, Ogura Y, et al. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 2009;206:79-87. [Crossref] [PubMed]
- Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 2010;11:404-10. [Crossref] [PubMed]
- Lee S, Ishitsuka A, Noguchi M, et al. Influenza restriction factor MxA functions as inflammasome sensor in the respiratory epithelium. Sci Immunol 2019; [Crossref] [PubMed]
- Meunier E, Broz P. Interferon-inducible GTPases in cell autonomous and innate immunity. Cell Microbiol 2016;18:168-80. [Crossref] [PubMed]
- Meunier E, Broz P. Interferon-induced guanylate-binding proteins promote cytosolic lipopolysaccharide detection by caspase-11. DNA Cell Biol 2015;34:1-5. [Crossref] [PubMed]
- Strandwitz P, Kim KH, Terekhova D, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 2019;4:396-403. [Crossref] [PubMed]
- Hagan T, Cortese M, Rouphael N, et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019;178:1313-1328.e13. [Crossref] [PubMed]
Cite this article as: Planès R, Goujon C. The surprising importance of gut microbiota in tackling flu infection. Biotarget 2020;4:3.


