
Suppression of the dynamic interaction of estrogen receptor with chromatin is critical for therapeutic ligands to repress ER-mediated transcription activities
Introduction
Breast cancer (BC) is the second leading cause of cancer deaths in women with annual new cases and fatality of 1.7 million and 500,000 respectively (1). Estrogen receptor-positive (ER+) BCs constitute 75% of the cases, and contribute to approximately 50% BC fatality (1). ERα plays critical roles in BC progression in part via transactivating Myc, cyclin D1, vascular endothelial growth factor, and other important oncogenic factors (2,3). Targeting ERα remains the standard of care in ER+ BCs; endocrine therapy (ET) is likely the most successful targeted cancer therapies. Adjuvant tamoxifen decreases mortality and recurrence by 31% and 50% respectively (4,5). The current toolbox of ET includes estrogen biosynthesis inhibitors (aromatase inhibitors, AIs) and therapeutic ligands; the latter consists of selective estrogen modulators (SERMs, like tamoxifen) and fulvestrant, a selective estrogen down-regulator (SERD). Although ET is clearly beneficial and with multiple options, ER+ BCs remains a major cause of BC mortality because of resistance. While resistance to ET (ETR) is mediated by complex mechanisms, persistent ER signaling under ET is a major attributor to the resistance; loss of ERα was reported in 17–28% of relapse BCs (6-8). The contributions of ERα in relapse BCs underlies multiple rounds of ET using alternative endocrine treatment. For instance, approximately 20% of relapse ER+ BCs following tamoxifen treatment are sensitive to AI and fulvestrant (9,10) and approximately 40% of recurrent ER+ BCs have mutations in ERα (11). Collectively, evidence supports an important role of persistent ERα function in ETR development.
The above situation also outlines a clear need to more effectively target ERα. Tamoxifen possesses partial agonist activities. In comparison, fulvestrant is a pure antagonist and thus a more potent antiestrogen, which is attributable to its action of inducing ERα degradation. However, the clinical application of fulvestrant is limited because of its poor solubility and intramuscular route of administration (12,13). This status underlies the current interest in developing new SERDs with improved pharmacokinetic properties for oral administration. Several of these SERDs have been developed and entered clinical trials, including GDC-0810 (multicenter phase Ia/IIa: NCT01823835), AZD9496 (phase I: NCT02248090; an open-label randomized multicenter trial: NCT03236974), RAD1901 (Elacetrant; phase III: NCT03778931; EudraCT 2018-002990-24), GDC-0927 (NCT02316509), and others.
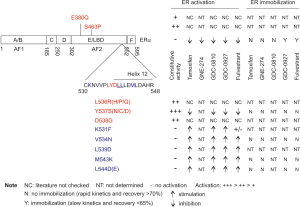
To develop effective antiestrogen ligands, a deep understanding of the mechanisms utilized by the current therapeutic ligands will provide a framework to guide this effort. ERα has been extensively investigated (the number of articles listed in PubMed under “estrogen receptor alpha”: n=20,816) following the cloning of human ERα from MCF7 cells in 1986 (14). ERα is a member of the nuclear receptor superfamily; it consists of 6 domains A-F with domain C and E as the DNA binding and the ligand-binding domain (LBD) respectively (Figure 1); the two activation functions are AF1 (domains A and B) and AF2 within the E domain (Figure 1) (16). The LBD motif is composed of 12 helices; reposition of the transactivation helix or helix 12 (h12) upon ligand binding defines ERα transcription activity. Binding of estradiol (E2) to LBD induces h12 to fold back, allowing h12 together with h3-h5 to form the co-activator binding groove (CBG) for co-activator recruitment (17). Binding of tamoxifen repositions h12 to the CBG space via association with h3-h4, preventing CBG formation (11,18). This antagonistic action can be reversed with mutations of two residues, L543A and L544A, in h12 of mouse ERα (mERα); binding of tamoxifen and fulvestrant but not E2 activates mERα (L543A, L544A) (16). Both L543 and L544 are conserved in the h12 of human ERα (L539 and L540) (Figure 1) (19). The L540Q mutation has been identified in a metastasis ER+ tumor after 5-year tamoxifen therapy (11). Similar to mERα (L543A, L544A), hERα (L540Q) is activated by antiestrogens tamoxifen, RU54876, and ICI164384 (an analogue of fulvestrant/ICI182780) (20). This knowledge together with the different efficacies of tamoxifen (partial agonist) and fulvestrant (full agonist) as antiestrogens suggests complex mechanisms underlying the antagonizing actions of antiestrogens.
To gain insights on these mechanisms, Guan et al. have reported an elegant and comprehensive study on a set of therapeutic ligands: tamoxifen, fulvestrant, some newly developed SERDs with better pharmacokinetic properties (GDC-0810, AZD9496, and GDC-0927), and GNE-274 which shares structural similarities with GDC-0927 without causing ERα degradation (15); the composition of this set of ligands contains ERα degraders (fulvestrant, GDC-0810, AZD9496, and GDC-0927) and non-ERα degraders (tamoxifen and GNE-274).
Associations of therapeutic ligands’ antiestrogen activities with their abilities to induce ERα degradation
Guan et al. started their research by examining the impact of tamoxifen, GNE-274, GDC-0810, AZD9496, GDC-0927, and fulvestrant on the proliferation of 6 ERα-positive and HER2-negative lines MCF7, MB-134 (MDA-MB-134-VI), HCC1500, EFM-19, CAMA-1, and T47D in vitro. Fulvestrant and GDC-0927 are more potent ERα degraders and inhibitors of cell proliferation; in MCF7, MB-134, and EFM-19 cells, ERα degraders (fulvestrant, GDC-0810, AZD9496, and GDC-0927) achieved higher levels of maximal inhibition on cell proliferation than tamoxifen and GNE-274. In vivo, GDC-0927 is more potent to inhibit E2-dependent growth of patient-derived ER+ xenograft (PDX) HCI-013 and HCI-011 compared to GDC-0810, consistent with the former inducing ERα degradation more effectively. Nonetheless, both antiestrogens displayed comparable inhibition of MCF7 xenograft growth (15). Additionally, RNA-seq analyses of gene expression of 8 ERα+ lines (the above 6 lines plus two HER2+ lines BT-474 and MDA-MB-340/MB-330) revealed the antagonistic activities (suppression of ERα-regulated gene expression) in the order of fulvestrant > GDC-0927 > GDC-0810 > tamoxifen and GNE-274. In both HCI-013 and HCI-011 PDXs, GDC-0927 exhibits superiority to GDC-0810 in suppression of ERα-regulated gene expression. These observations support a positive correlation between the level of antagonistic activities of these therapeutic ligands and their ability to induce ERα degradation.
Induction of chromatin recruitment of ERα by therapeutic ligands
SERDs as antiestrogens are widely regarded to be attributable to their properties of inducing ERα degradation, a concept that is in line with the authors’ functional studies. Nonetheless, the degradation property of fulvestrant and its antagonist actions can be uncoupled in vitro (21). It has been well established that the proper reposition of h12 upon agonist binding is critical for LBD to form an active AF2 in part through formation of CBG, allowing co-activator recruitment and that antagonist binding induces the occupation of h12 to CBG position, preventing co-activator association (18). With this knowledge, Guan et al. examined the displacement of 154 co-factor peptides which potentially contribute to nuclear receptor signaling (22), and observed a similar profile of co-factor competition among tamoxifen, GNE-274, AZD9496, GDC-0810, GDC-0927, and fulvestrant (15). This finding implies a common mechanism in the suppression of ERα transcription functions, instead of ERα degradation.
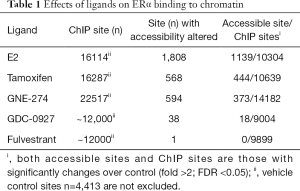
Ligand-binding initiates the recruitment of ERα to chromatin, a step required for ERα to regulate transcription. Remarkably, both GNE-274 and GDC-0927 induce a rapid ERα recruitment in MCF7 cells following 10 minute stimulation, despite only GDC-0927 leads to ERα degradation; at 45 minutes, all antagonists (tamoxifen, GNE-274, AZD9496, GDC-0810, GDC-0927, and fulvestrant) cause comparable or higher levels of ERα association with chromatin compared to E2–treated MCF7 cells. Furthermore, a systemic examination of ligand-induced ERα binding to chromatin using ChIP-seq revealed robust increases in ERα binding sites from 4,413 in vehicle-treated MCF7 cells to 12,000 (GDC-0927 or fulvestrant), 16,114 (E2), 16,287 (tamoxifen), and 22,517 (GNE-274) in cells with the indicated treatment (Table 1). Importantly, 57.9% (9,326/16,114) of E2-induced binding sites overlap with GNE-274 or GDC-0927; an overall large proportion of E2-induced binding sites are bound by ERα when associated with these therapeutic ligands. Collectively, therapeutic ligands are capable of inducing ERα to bind to estrogen response element (ERE) sites in a manner comparable to E2, which is independent of their ability to induce ERα degradation.
Full table
Differential impacts on chromatin accessibility by ERα-liganded with agonists or antagonists
The comparable binding of ERα-E2 and ERα-antagonist to chromatin raises the issue whether both ERα-ligand complexes affect chromatin configuration similarly. To address this, the authors performed another comprehensive research: Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) to profile open chromatin regions. In MCF7 cells treated for 45 minutes with E2, tamoxifen, GNE-274, GDC-0927, or fulvestrant, the number of sites with chromatin accessibility altered was 1,808, 568, 594, 38, or 1 with predominant changes for increases in accessibility (Table 1) (15). Furthermore, among the ERα binding sites detected by ChIP in MCF7 cells treated with E2, tamoxifen, GNE-274, GDC-0927, or fulvestrant, sites with significantly altered accessibility (fold change >2; FDR <0.05) are 1139/10304 (accessible site/ChIP sites), 444/10639, 373/14182, 18/9004, or 0/9899 (Table 1) (15). These altered accessible sites are enriched for ERE motif, and are associated with transcription activity at least for some ERα target genes (ADORA1, PGR, RET, AGR3, and FKBP4) in MCF7 cells treated with E2, tamoxifen, GNE-274, GDC-0810, GDC-0927, or fulvestrant (15). Taken together, data demonstrate that the inaccessibility of chromatin contributes to the antagonistic potential of individual therapeutic ligands examined. These observations are in accordance with the current knowledge that binding of antagonists prevents co-activator association (18).
Impairment of the dynamics of ERα-chromatin association by fulvestrant and GDC-0927
Narrowing down the differences between ERα-agonist and ERα-antagonist to their unique association with chromatin, the authors examined their dynamic association with chromatin, which is an emerging feature of transcription factors (23). MCF7 cells were engineered to stably express mNeon-tagged ERα; the ectopic protein performs similar as the endogenous counterpart in terms of ligand-induced degradation. These cells were treated with individual ligands for 45 minutes prior to photobleaching, followed by monitoring the recovery of fluorescence signals within 60 seconds. The recovery is in a rapid kinetics with a final recovery of 70–80% in cells treated with tamoxifen, GNE-274, GDC-0810, or AZD9496 in comparison to a much slower and less recovery (60–65%) in cell treated with either GDC-0927 or fulvestrant (15). Guan et al. concluded that immobilization of ERα by fulvestrant and GDC-0927 is a causation of ER degradation.
To show ERα mobility being relevant to ER turn over and its transcription activity, Guan et al. aimed to find ERα mutants that reverse antagonists into agonists with concurrent improvements of ERα mobility. The mutations of mERα (L543A, L544A) (16) and hERα (L540Q) reverse antagonists into agonists (11,20); mutations of Y537S, D538G, L536, S463P, and E380Q were detected in recurrent ER+ BCs (Figure 1); and h12 plays critical roles in forming AF2 (11). With this knowledge, the authors have performed a systemic mutagenesis in 20 residues, including E380, S463, R503, and amino acids 530–546 (Figure 1). Along with the re-discovery of Y537S, a set of mutants were identified from mutations in K531F, V534N, and h12 residues: L539D (similar to L543A in mERα), M543K, and L544D/E (Figure 1) (15). Fulvestrant acts as an agonist to V534N, L539D, and M534K (Figure 1) without inducing their degradation and compromising their mobility (15). Other therapeutic ligands (tamoxifen, GNE-274, and GDC-0927) also activate L539D (Figure 1). The agonist activities of tamoxifen, GNE-274, GDC-0927, and fulvestrant towards these mutants were not derived from AF2, as L539D does not clearly recruit co-activator peptides (n=154) in the presence of E2, tamoxifen, GNE-274, GDC-0810, GDC-0927, or fulvestrant. However, inactivation mutation of AF1 in hERα (L539D) prevents all therapeutic ligands (tamoxifen, GNE-274, GDC-0927, and fulvestrant) from inducing L539D activation, indicating transcription activities being attributable to AF1 (15).
Perspectives
By taking multiple systematic approaches (RNA-seq, ChIP-seq, ATAC-seq, and mutational screen), this research reveals that the antiestrogen functions of SERDs are not attributable to their ability of inducing ERα degradation, neither to preventing ERα from binding to ERE sites. These observations are in accordance with (I) the formation of the same dimers by binding of either agonists or antagonist and (II) ERα dimers binding DNA (17). Nonetheless, antagonist-liganded ERα is unable to make these ERE motifs accessible for transcription activities. This knowledge may provide a framework to develop more effective ERα antagonists, which may not focus on their ability to cause ERα degradation. In this regard, it will be interesting to investigate H3B-5942 actions of antagonist. H3B-5942 is a newly developed selective estrogen receptor covalent antagonist (SERCA) which causes ERα degradation through covalent targeting C530 (24). Nonetheless, the conclusions of this article require further investigations, as both ChIP-seq and ATAC-seq were essentially carried out in a single MCF7 cell line.
While the ERα mobility studies coupled with mutagenesis screening support an association of ERα immobilization with fulvestrant- and GDC-0927-derived ERα degradation, whether immobilization contributes to ERα turn over and suppression of ERα transcription regulation remains unclear. In the mNeon-tagged ERα stable lines, GDC-0810 displayed a comparable efficiency in inducing ERα degradation to GDC-0927 without clear immobilization of ERα (Figure 1). In parental MCF7 cells, GDC-0927 does not apparently lead to ERα degradation but does so in mNeon-tagged ERα stable line. Furthermore, tamoxifen, GNE-274, GDC-0810, and AZD9496 are all largely antagonists without apparently affecting ERα motility. Lastly, ICI164384 was reported to relocate h12 to a position without association with other LBD domains in ERβ, which may destabilize ERβ (25). The possibility for a similar mechanism in fulvestrant-liganded ERα should be considered. Clearly, the necessity of ERα mobility needs to be further elucidated, as well as the immobilization effects of fulvestrant and GDC-0927. For instance, how will binding of fulvestrant or GDC-0927 to the loss of function mutant (AF1mut.L539D) affect the ERα mobility?
Mutation of L511R in mouse ERα (L543A, L544A) renders tamoxifen and fulvestrant to be incapable of inducing mERα dimerization and activation (16); the residue is conserved (L507) in hERα. It will be interesting to examine the performance of hERα (L507; L539D) with the set of therapeutic ligands studied by Guan et al. As formation of dimers via a ligand binding sets the motion of ERα binding to DNA, can the mERα (L511R, L543A, L544A) structural information or hERα (L507; L539D) be explored for developing antagonists to prevent ERα dimerization?
Acknowledgments
Funding: This work is supported by a grant from Canadian Cancer Society (grant #: 319412) to D Tang.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Siegel RL, Miller KD, Jemal A. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Musgrove EA, Caldon CE, Barraclough J, et al. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011;11:558-72. [Crossref] [PubMed]
- Rodriguez D, Ramkairsingh M, Lin X, et al. The Central Contributions of Breast Cancer Stem Cells in Developing Resistance to Endocrine Therapy in Estrogen Receptor (ER)-Positive Breast Cancer. Cancers (Basel) 2019;11. [Crossref] [PubMed]
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005;365:1687-717. [Crossref] [PubMed]
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009;9:631-43. [Crossref] [PubMed]
- Johnston SR, Saccani-Jotti G, Smith IE, et al. Changes in estrogen receptor, progesterone receptor, and pS2 expression in tamoxifen-resistant human breast cancer. Cancer Res 1995;55:3331-8. [PubMed]
- Gutierrez MC, Detre S, Johnston S, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol 2005;23:2469-76. [Crossref] [PubMed]
- Dowsett M. Overexpression of HER-2 as a resistance mechanism to hormonal therapy for breast cancer. Endocr Relat Cancer 2001;8:191-5. [Crossref] [PubMed]
- Osborne CK, Pippen J, Jones SE, et al. Double-blind, randomized trial comparing the efficacy and tolerability of fulvestrant versus anastrozole in postmenopausal women with advanced breast cancer progressing on prior endocrine therapy: results of a North American trial. J Clin Oncol 2002;20:3386-95. [Crossref] [PubMed]
- Howell A, Pippen J, Elledge RM, et al. Fulvestrant versus anastrozole for the treatment of advanced breast carcinoma: a prospectively planned combined survival analysis of two multicenter trials. Cancer 2005;104:236-9. [Crossref] [PubMed]
- Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, et al. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer 2018;18:377-88. [Crossref] [PubMed]
- Soleja M, Raj GV, Unni N. An evaluation of fulvestrant for the treatment of metastatic breast cancer. Expert Opin Pharmacother 2019;20:1819-29. [Crossref] [PubMed]
- Robertson JF. Fulvestrant (Faslodex) -- how to make a good drug better. Oncologist 2007;12:774-84. [Crossref] [PubMed]
- Greene GL, Gilna P, Waterfield M, et al. Sequence and expression of human estrogen receptor complementary DNA. Science 1986;231:1150-4. [Crossref] [PubMed]
- Guan J, Zhou W, Hafner M, et al. Therapeutic Ligands Antagonize Estrogen Receptor Function by Impairing Its Mobility. Cell 2019;178:949-63.e18. [Crossref] [PubMed]
- Arao Y, Korach KS. Transactivation Function-1-Mediated Partial Agonist Activity of Selective Estrogen Receptor Modulator Requires Homo-Dimerization of the Estrogen Receptor alpha Ligand Binding Domain. Int J Mol Sci 2019;20. [Crossref] [PubMed]
- Ruff M, Gangloff M, Wurtz JM, et al. Estrogen receptor transcription and transactivation: Structure-function relationship in DNA- and ligand-binding domains of estrogen receptors. Breast Cancer Res 2000;2:353-9. [Crossref] [PubMed]
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 2000;21:381-8. [Crossref] [PubMed]
- Danielian PS, White R, Lees JA, et al. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J 1992;11:1025-33. [Crossref] [PubMed]
- Montano MM, Ekena K, Krueger KD, et al. Human estrogen receptor ligand activity inversion mutants: receptors that interpret antiestrogens as estrogens and estrogens as antiestrogens and discriminate among different antiestrogens. Mol Endocrinol 1996;10:230-42. [PubMed]
- Wardell SE, Marks JR, McDonnell DP. The turnover of estrogen receptor alpha by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochem Pharmacol 2011;82:122-30. [Crossref] [PubMed]
- Wang S, Houtman R, Melchers D, et al. A 155-plex high-throughput in vitro coregulator binding assay for (anti-)estrogenicity testing evaluated with 23 reference compounds. ALTEX 2013;30:145-57. [Crossref] [PubMed]
- Liu Z, Tjian R. Visualizing transcription factor dynamics in living cells. J Cell Biol 2018;217:1181-91. [Crossref] [PubMed]
- Puyang X, Furman C, Zheng GZ, et al. Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ERalpha(WT) and ERalpha(MUT) Breast Cancer. Cancer Discov 2018;8:1176-93. [Crossref] [PubMed]
- Pike AC, Brzozowski AM, Walton J, et al. Structural insights into the mode of action of a pure antiestrogen. Structure 2001;9:145-53. [Crossref] [PubMed]
Cite this article as: Chow MJ, Peng J, Su Y, Tang D. Suppression of the dynamic interaction of estrogen receptor with chromatin is critical for therapeutic ligands to repress ER-mediated transcription activities. Biotarget 2020;4:1.


