
Fragmentation patterns of circulating cell-free DNA demonstrate biomarker potential for human cancers
Introduction
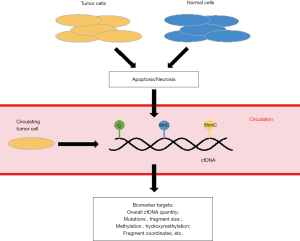
The often-dismal clinical outcomes for most cancers are due to the fact that cancer patients diagnosed at advanced stages would miss the opportunities for curative treatments or effective treatments no longer available for these patients (1). Therefore, lacking clinically proven and convenient tools for early detection of cancers has been one major contributor to the poor clinical outcomes for many human cancers. In particular, considering their non-invasiveness and clinical relevance, the past decade has witnessed significant efforts of exploring patient-derived biofluids (i.e., liquid biopsy) in cancer biomarker discovery. Of great interest to the research community has been the potential utility of circulating cell-free DNA (cfDNA) in the blood as a non-invasive biomarker target. Specifically, cfDNA in patient-derived blood contains DNA fragments released from the tumor cells or from such cellular mechanisms as apoptosis and necrosis (Figure 1). Importantly, patient-derived cfDNA contains genetic and epigenetic information (e.g., mutations, cancer-associated cytosine modifications) from the tumor origin, which has been exploited to identify biomarkers for various human cancers, showing tremendous promise as a non-invasive biomarker target (3-6).
In contrast to previous studies that target genetic or epigenetic markers in cfDNA, recently, Cristiano and coworkers reported a new approach that exploits the cfDNA fragmentation patterns for cancer diagnosis (2). Their approach identified altered fragmentation profiles across the genomes in 236 patients with a variety of cancers, compared to 245 healthy individuals whose fragmentation patterns reflected nucleosomal patterns of white blood cells. Findings from Cristiano et al. opened up new opportunities of using cfDNA as a non-invasive tool for cancer biomarker discovery by targeting a novel molecular feature in patient-derived cfDNA, showing the promise of more innovative applications of cfDNA in precision oncology.
Liquid biopsy as a promising target for cancer biomarker discovery
For cancer biomarker discovery, tissue biopsy has been the “gold-standard”, because tissue samples are believed to reflect molecular features of tumors, e.g., genetic and epigenetic features, as well as cancer type specificity (4). However, due to the invasive nature, the procedure of obtaining tissue biopsy can be accompanied by clinical risks such as bleeding and infection (7). In addition, owing to the intra-tumor heterogeneity, a hallmark of human cancers, single site biopsies may only provide limited information of the whole tumor (8). Thus, multi-site tissue biopsies, which could be difficult to obtain because of tumor accessibility (9,10), may be required for accurate pathological confirmation and diagnosis. Furthermore, continuous monitoring of disease progression is crucial during treatment (e.g., surveillance of therapeutic response). However, it may not be always clinically feasible for routine tumor biopsies along the treatment course for patients (11).
In contrast, liquid biopsy, such as cfDNA that can be isolated from the plasma of peripheral blood, is emerging as a non-invasive and clinically convenient tool for cancer biomarker discovery, with potential clinical applications in early detection, diagnosis, and prognosis. The majority of previous studies have explored genetic or epigenetic markers in cfDNA for cancer biomarker discovery, because cfDNA is believed to contain genetic and epigenetic information from the tumor origin. Recent cfDNA-based studies suggested that cancer-specific genetic alterations could be used for cancer early detection and disease surveillance using this non-invasive target (12-14). For example, whole-genome sequencing (WGS) of cfDNA has been used to identify chromosomal or genetic abnormalities associated with cancer patients (15-18). However, detecting such chromosomal alterations or single genetic alterations may be extremely challenging owing to the small number of abnormal chromosomal changes, the heterogeneity of tumors, and the small amount of tumor-derived cfDNA in the plasma (13). Based on the hypothesis that detection of a larger number of alterations in cfDNA may be more sensitive for cancer detection, a recent research by Cristiano et al. developed an approach called the “DNA evaluation of fragments for early interception” (DELFI), which aimed to detect the abnormalities of cfDNA fragmentation profiles at genome-wide level for various cancer patients and use this novel molecular feature of cfDNA in cancer biomarker discovery (2).
Cancer detection taking advantage of aberrant cfDNA fragmentation profiles in the blood
Technically, Cristiano et al.’s method was based on the low-coverage WGS of cfDNA isolated from the plasma. Coverage and size distribution of cfDNA fragments in non-overlapping 5-megabase (Mb) windows that covered the genome were examined for healthy individuals and cancer patients. The genome-wide fragmentation pattern from an individual was categorized for cancer diagnosis and tumor tissue of origin identification using machine learning. They have found that the length of cfDNA fragmentation derived from cancer patients were more variable than that that from healthy individuals (P<0.001). Thus, they focused on the fragmentation size of cfDNA in their analysis. Fractions of small cfDNA fragments (100–150 bp) to larger cfDNA fragments (151–220 bp) in 504 slide windows of 5 Mb across the genome in a position-dependent manner were obtained for each sample and defined as the genome-wide fragmentation profiles. Healthy individuals showed similar genome-wide profiles, which were highly correlated with lymphocyte nucleosomal DNA fragmentation profiles and nucleosome distances, suggesting that fragmentation patterns of cfDNA derived from healthy individuals were likely the result of nucleosomal DNA patterns that reflected the chromatin structure of normal blood cells. In contrast, patients with cancer showed several distinct genomic differences from healthy individuals, with variable fragment sizes at different regions. Interestingly, the fragmentation profiles in patient-derived cfDNA were significantly correlated with mutant allele fractions of the levels of EGFR or ERBB2 for patients undergoing anti-EGFR or anti-ERBB2 treatment, suggesting its potential as a cancer management tool.
By integrating the copy number changes from chromosomal arm features and mitochondrial copy number changes into the DELFI approach, Cristiano et al. obtained a score that could be used for overall cancer detection, achieving 80% sensitivity, 95% specificity, and an AUC (area under the curve) of 0.94 (95% CI: 0.92–0.96), with an AUC of at least 0.92 for each tumor stage. The AUCs for distinguishing seven cancers (e.g., pancreatic, breast, bile duct, colorectal, gastric, lung, and ovarian cancer) from healthy individuals ranged from 0.86 to 1. Additionally, the performance of the DELFI approach was further improved by combining with mutation detection in cfDNA.
Moreover, since cfDNA can be originated from tumor tissues, reflecting the genetic and epigenetic alterations of tumor tissues (19), cfDNA fragmentation profiles may reveal regional differences between tumors. Using machine learning, Cristiano et al. were able to estimate the tissue of origin of circulating tumor DNA with 75% (95% CI: 69–81%) accuracy, suggesting its potential application in a pan-cancer detection algorithm.
Conclusions
Fragmentation profiling in circulating cfDNA introduced in Cristiano et al. represented one of the latest efforts of exploiting molecular features in cfDNA for cancer biomarker discovery and provided an alternative non-invasive diagnostic tool that could improve early detection of human cancers, thus showing its potential as a screening and management tool for precision oncology. Considering its potential clinical impact, we would recommend several directions for future studies. Firstly, extensive validation using clinical samples from patients with confounding diseases (e.g., liver cancer vs. liver cirrhosis) will help establish the sensitivity and clinical utility of the DELFI approach. Secondly, it will be of interest for future studies of larger sample sizes to comprehensively evaluate whether the DELFI approach is informative for tumor stages and tumor malignancy, or has prognostic value. Thirdly, by focusing on the fragmentation patterns, the DELFI approach understandably missed opportunities to utilize other important molecular features of cfDNA (e.g., epigenetic markers). For example, considering recent progress in exploiting novel epigenetic markers in cfDNA, such as 5-hydroxymethylcytosines, in biomarker discovery for human cancers (3,20,21), integrating both the fragmentation patterns and epigenetic features of cfDNA could possibly enhance our understanding of the underlying mechanisms of molecular alterations in the cancer genome and epigenome, as well as improve the detection performance. In summary, Cristiano et al. provided exciting new progress in this emerging area with potentially tremendous impact on clinical oncology. Though promising, applying this novel approach in a clinical setting is still challenging and has a long way to go. Extensive validation and trials will be needed for the ultimate application of the DELFI approach, possible in combination with other approaches, in the clinic. We are closer to the goal of precision oncology by another important step.
Acknowledgments
Funding: This work was supported partially by grants from the National Institutes of Health (to WZ) (R01CA223662, R21MD011439, and U01CA217078).
Footnote
Conflicts of Interest: W Zhang is a shareholder of Shanghai Epican Genetech Co. Ltd., which develops epigenetic cancer biomarkers. Z Zhang has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385-9. [Crossref] [PubMed]
- Cai J, Chen L, Zhang Z, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Zeng C, Stroup EK, Zhang Z, et al. Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun (Lond) 2019;39:12. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579-83. [Crossref] [PubMed]
- Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64:876-92. [Crossref] [PubMed]
- Sabaawy HE. Genetic Heterogeneity and Clonal Evolution of Tumor Cells and their Impact on Precision Cancer Medicine. J Leuk (Los Angel) 2013;1:1000124. [Crossref] [PubMed]
- Ilie M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res 2016;5:420-3. [Crossref] [PubMed]
- Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225-33. [Crossref] [PubMed]
- Khan KH, Cunningham D, Werner B, et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer Discov 2018;8:1270-85. [Crossref] [PubMed]
- Park G, Park JK, Son DS, et al. Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients. Sci Rep 2018;8:11631. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017. [Crossref] [PubMed]
- Cohen JD, Li L, Wang YX, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Leary RJ, Kinde I, Diehl F, et al. Development of Personalized Tumor Biomarkers Using Massively Parallel Sequencing. Sci Transl Med 2010;2:20ra14. [Crossref] [PubMed]
- Leary RJ, Sausen M, Kinde I, et al. Detection of Chromosomal Alterations in the Circulation of Cancer Patients with Whole-Genome Sequencing. Sci Transl Med 2012;4:162ra154. [Crossref] [PubMed]
- Ossandon MR, Agrawal L, Bernhard EJ, et al. Circulating Tumor DNA Assays in Clinical Cancer Research. J Natl Cancer Inst 2018;110:929-34. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 2017. [Crossref] [PubMed]
- Song CX, Yin S, Ma L, et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 2017;27:1231-42. [Crossref] [PubMed]
- Li W, Zhang X, Lu X, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 2017;27:1243-57. [Crossref] [PubMed]
Cite this article as: Zhang Z, Zhang W. Fragmentation patterns of circulating cell-free DNA demonstrate biomarker potential for human cancers. Biotarget 2019;3:16.


