
Long noncoding RNA MALAT1 and cancer metastasis
Since Malat1 was identified as a prognostic marker for poor clinical outcomes in non-small cell lung cancer (NSCLC) patients, number of Malat1 studies have revealed its roles in human cancers (1,2). Recently, we demonstrated unexpected Malat1 function in suppressing breast cancer metastasis using genetic mouse and cell line models of breast cancer (3). Here, I review the controversies in Malat1 studies conducted by ourselves and others by comparing research designs and interpreting the results.
Previously, Malat1 was shown to co-localize with nuclear speckles enriched in pre-mRNA splicing factors. In vitro siRNA study revealed that Malat1 interacts with splicing factors in the nucleus and regulates pre-mRNA splicing (4). Malat1 genetic knockout mice were generated from independent research groups and exhibited no apparent phenotypic abnormalities (5,6). Surprisingly, unlike in vitro results, there were no significant changes in alternative splicing or global gene expression due to genetic loss of Malat1 (3,6).
Arun et al. (7) generated Malat1 knockout mice by deleting 3 kb genomic locus spanning both upstream and downstream of transcription start site and crossed them to Polyomavirus middle T antigen (PyMT)-induced mouse mammary tumor model. Although Malat1 loss did not substantially alter primary tumorigenesis, mammary tumors of Malat1−/− PyMT mice exhibited cystic phenotype compared to Malat1+/+ PyMT animals. Furthermore, lung metastasis was significantly suppressed by Malat1 loss. In addition to genetic ablation of Malat1, PyMT mice were treated with antisense oligonucleotides (ASOs). Compared to complete genetic Malat1 knockout, ASOs led to ~60% reduction in Malat1 levels but caused stronger suppression in mammary tumor growth and comparable inhibition in lung metastasis. Targeting Malat1 with less-effective ASOs demonstrated more evident tumor-suppressive outcome, which obviously raises a question of off-target effects of ASOs.
We utilized Malat1 mouse knockout model generated by targeted insertional inactivation (5) and crossed them to PyMT mice (3). Unexpectedly, however, we observed dramatic induction of lung metastasis in Malat1−/− PyMT mice while there was no noticeable difference in mammary tumors compared to Malat1+/+ PyMT mice; cystic and high-grade tumor areas were similarly found in both groups. Instead, we could see consistent increase in circulating tumor cells from the peripheral blood of Malat1−/− PyMT mice, which implies that Malat1 loss affects certain stage of metastatic process. This surprising result against the current dominant paradigm of Malat1’s pro-metastatic function prompted us to restore Malat1 in Malat1−/− PyMT mice using transgenic animals to determine if metastasis-promoting function is attributed to Malat1 loss. When Malat1 was re-expressed, metastatic phenotype was dramatically reversed. Furthermore, considering higher Malat1 levels in Malat1-restored PyMT mice compared to Malat1+/+ PyMT mice, Malat1-re-expression resulted in less metastasis than Malat1+/+, which implies dose-dependent effect of Malat1 in lung metastasis suppression. Using more aggressive PyMT mice on FVB/N background, we overexpressed Malat1 by crossing them to Malat1 transgenic mice on FVB/N and observed that lung metastasis was significantly suppressed by transgenic overexpression of Malat1.
In addition, we inactivated MALAT1 in MDA-MB-231 human breast cancer cells using CRISPR-Cas9 by deleting ~650 bp in 5' genomic locus of MALAT1. MALAT1 expression was completely abrogated unlike siRNAs, shRNAs or ASOs which partially suppressed nuclear-enriched RNAs. MALAT1 knockout dramatically elevated metastatic potential, which were reversed by re-expression of Malat1. Furthermore, when Malat1 was overexpressed in highly metastatic 4T1 mouse mammary tumor cells as well as lung metastatic subline of MDA-MB-231 (LM2), metastasis in both cell lines was significantly suppressed.
What possibly causes the substantial difference in studying the same lncRNA Malat1? There is one example that inactivating a lncRNA in mice by different strategies exhibited opposite phenotypes. LncRNA Haunt expression was inactivated by either inserting transcription stop signal in the downstream of transcription start site or deleting various lengths of genomic locus of Haunt (2.3–58 kb) (8). When transcription stop signal was inserted or short genomic region was deleted, expression of HoxA gene cluster located in the downstream of Haunt was significantly elevated. However, the effect was attenuated when longer genomic regions were deleted. This turned out that Haunt genomic locus contains enhancer sequence regulating HoxA expression, so minimal deletion/insertional inactivation and long genomic deletion demonstrated opposite gene expression patterns. Moreover, Haunt re-expression failed to rescue knockout phenotype driven by deletion of longer genomic locus. This study implicates that deleting genomic locus to inactivate the target lncRNA may disrupt cis regulatory elements, and it is essential to restore the lncRNA expression in order to examine if the phenotype driven by lncRNA inactivation is rescued. In fact, Malat1 inactivation by genomic deletion resulted in significant transcriptional upregulation of its neighbor genes in cis (6) while insertional inactivation of Malat1 did not (1), which suggests that deleted genomic region may contain cis-regulatory sequences for Malat1-neighboring genes.
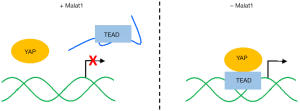
To elucidate molecular mechanism by which Malat1 suppresses breast cancer metastasis, we searched for Malat1-interacting proteins in mouse mammary tumors and identified Tead family members, the oncogenic and pro-metastatic transcription factors (Tead1–4). In the subsequent analyses, we found that Tead binding sites are distributed throughout Malat1, and Malat1 could inactivate Tead proteins by sequestering them from interacting with Yap coactivator (Figure 1). However, any of previous Malat1 knockout mouse studies did not report abnormalities in Malat1−/− mice due to Tead-Yap hyperactivation (5,6). In human mammary tissue, for example, YAP is highly enriched in the nuclei of breast tumor cells while localized more in the cytoplasm of normal mammary cells (9). Notably, nuclear localization of Yap was prominent in the mammary tumor cells of PyMT mice in our unpublished data. This suggests that Malat1 inactivation needs to be combined with nuclear translocation of Yap to interact with Tead proteins, which fully enhances their transcriptional potential. For this reason, cooperation model of Malat1 deficiency and nuclear Yap warrants future studies.
In conclusion, most Malat1 research including in vitro and in vivo studies has not been confirmed by rescue experiments which we rigorously conducted recently (3). Especially for finding therapeutic application of RNA interference or ASOs targeting lncRNA MALAT1 in human cancers, it is critical to exclude any possibility of their off-target effects by performing rescue experiments with accurate controls (10).
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Bi S, Wang C, Li Y, et al. LncRNA-MALAT1-mediated Axl promotes cell invasion and migration in human neuroblastoma. Tumour Biol 2017;39:1010428317699796. [Crossref] [PubMed]
- Kwok ZH, Roche V, Chew XH, et al. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int J Cancer 2018;143:668-78. [Crossref] [PubMed]
- Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 2018;50:1705-15. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
- Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012;18:1487-99. [Crossref] [PubMed]
- Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2012;2:111-23. [Crossref] [PubMed]
- Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016;30:34-51. [Crossref] [PubMed]
- Yin Y, Yan P, Lu J, et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell 2015;16:504-16. [Crossref] [PubMed]
- Steinhardt AA, Gayyed MF, Klein AP, et al. Expression of Yes-associated protein in common solid tumors. Hum Pathol 2008;39:1582-9. [Crossref] [PubMed]
- Kaelin WG Jr. Common pitfalls in preclinical cancer target validation. Nat Rev Cancer 2017;17:425-40. [Crossref] [PubMed]
Cite this article as: Kim J. Long noncoding RNA MALAT1 and cancer metastasis. Biotarget 2019;3:5.


