
Targeting the BECN1-BCL2 autophagy regulatory complex to promote longevity
Several studies in the past two decades established an evolutionarily conserved and tight relationship between autophagy and ageing. Model organisms such as the budding yeast Saccharomyces cerevisiae, the soil dueling worm Caenorhabditis elegans and the fruit fly Drosophila melanogaster, have been instrumental for the identification of key genes regulating a variety of pathophysiological processes and signaling, including ageing and associated pathologies (1). Together with fundamental studies in mammalian systems, model organisms also helped deciphering different hallmarks of ageing, defined as biological processes which become dysfunctional during ageing—often in a tissue specific manner—and whose genetic or pharmacological manipulation delay or accelerate the rate of ageing (2,3). Among these, an evolutionarily conserved one is protein homeostasis, which requires different intracellular molecular mechanisms including autophagy for the turnover of old, damaged or dysfunctional proteins. Indeed, the autophagic process (from bulk autophagy to more specialized forms of autophagy such as mitophagy and lipophagy) as well as the expression of autophagy regulatory genes decline with ageing. Alongside, seminal studies in different model organisms revealed that many autophagy regulatory genes are causally involved in the lifespan extension elicited by different genetic, dietary or pharmacological manipulations (4-7). These studies revealed the complex interaction between autophagy and other key biological homeostatic processes, such as lipid metabolism, mitochondrial function, neuronal activity and host defense, in regulating ageing and associated pathologies. Intriguingly, a growing body of evidence revealed that more and more genes regulating autophagy are in fact also involved in other biological processes ranging from inflammation to apoptosis (8). Moreover, thanks to their easy manipulability and short lifespan, model organisms allowed the screening and identification of several extrinsic interventions ranging from dietary and pharmacological compounds to small molecules, which extends lifespan by inducing the autophagic flux (4). They also helped to disclose the importance of tissue-, context- and age-specific effects in autophagy control of longevity. For instance, in the nematode C. elegans, inactivation of autophagy during development is deleterious and suppresses the life-extending effect of different longevity paradigms (1,9), while inactivation after the fertile period promotes health and lifespan extension (10).
Although studies in model organisms will continue to shed light on mechanisms regulating the different forms of autophagy and their activation in response to specific types of stress or metabolic changes, studies in mammals needs to be implemented in order to validate and propose autophagy as a therapeutic target for ageing and associated pathologies (11,12). Of note, despite our knowledge on the role of autophagy in the ageing process, definite evidence indicating that increased basal autophagy flux can promote healthy ageing in mammals was so far missing. The recent collaborative effort between the groups of Dr. Hu and Dr. Levine (both at University of Texas Southwestern Medical Center, Dallas, TX, USA) elegantly fills this gap in the field by providing the first evidence that genetic disruption of the Beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice by increasing basal autophagy (13).
Beclin 1 (BECN1) is a component of the class III phosphatidylinositol-3-OH kinase (PI3K) complex. Similar to yeast, also in mammals there exist at least two distinct PI3K complexes, termed complexes I and II (14). Both complexes contain the catalytic subunit PIK3C3, the regulatory subunit PIK3R4, and BECN1. BECN1 contains an N-terminal intrinsically disordered region, a BCL2 homology 3 (BH3) domain, a coiled-coil domain, and a C-terminal β-α repeated, autophagy-specific (BARA) domain (15). Via its coiled coil domain, BECN1 interacts with ATG14, which targets the PI3K complex I to the phagophore assembly site (PAS), where the biogenesis of autophagosomes is initiated (14). This PIK3C3-PIK3R4-BECN1-ATG14 core complex reveals a V-shaped architecture. This heterotetramer associates with a fifth subunit termed nuclear receptor binding factor 2 (NRBF2), and one NRBF2 dimer bridges two PIK3C3-PIK3R4-BECN1-ATG14 heterotetramers (14). The class III PI3K3 complex produces phosphatidylinositol 3-phosphate (PI3P), which then recruits additional autophagy-specific downstream factors, including proteins of the WIPI family and ZFYVE1 (16).
The PI3K complex I receives regulatory input via post-translational modifications and several protein-protein interactions, and BECN1 is at the center of this regulation (14,15). For example, BECN1 is phosphorylated by at least 8 kinases, both at stimulatory and inhibitory sites. These kinases are ULK1, MAPKAPK2/3, AMPK, DAPK1, ROCK1, STK4, AKT, and EGFR (14,15). BECN1-binding partners include AMBRA1 or BCL2 family members (14,15). In fact, BECN1 has originally been identified as BCL2-interacting protein by a yeast-two-hybrid screen (17). Subsequently, the functional relevance of this interaction has been reported. Next to its anti-apoptotic function, BCL2 can inhibit starvation-induced, BECN1-dependent autophagy (18). Also the BECN1-BCL2 interaction is regulated by multiple stimuli, including competitive binding, self-association, phosphorylation, or ubiquitination (Figure 1) (15,19). The BH3 domain is targeted by the kinases DAPK1, ROCK1 (both phosphorylating T119) and STK4 (phosphorylating T108), and phosphorylation either inhibits (phospho-T119) or promotes (phospho-T108) the association between BECN1 and BCL2 (21-23). Similarly, K117 in the BH3 domain can become ubiquitinated by TRAF6 (24), presumably leading to a reduced BECN1-BCL2 interaction. Finally, the dissociation of BECN1 from BCL2 can be promoted by MAPK8-mediated phosphorylation of BCL2, BH3-only proteins, or HMGB1-binding to BECN1 (15,20).
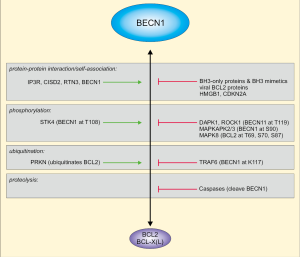
The modulation of the BECN1-BCL2 interaction is central for the work presented by Fernandez et al. (13). The authors make use of a mouse strain expressing a F121A variant of BECN1 (corresponding to F123A in human BECN1), which exhibits decreased binding to BCL2. In a previous work, it has been demonstrated that autophagy is hyperactivated in these mice, leading to the sequestration of amyloid oligomers and the prevention of Alzheimer’s disease progression (25). In the present work, Fernandez et al. investigated the effect of the genetic disruption of the BECN1-BCL2 complex on the health- and lifespan of these mice (13). The authors observe increased basal autophagy in these knock-in mice, and a sex-independent extension of lifespan. Furthermore, they reported decreased age-related renal and cardiac changes, and a decreased incidence of age-related spontaneous cancer. Of note, the decreased negative regulation of BECN1 in the knock-in mice was per se not sufficient to reverse the age-related decline in autophagic function. Finally, the authors investigated whether the known anti-ageing and pro-autophagic factor klotho exerts its activity by disrupting the BECN1-BCL2 interaction. This hypothesis was supported by their observation that klotho deficiency resulted in an increased association of BECN1 with BCL2. Of note, the F121A mutation reversed the premature ageing phenotypes—such as increased lethality, infertility or growth retardation—resulting from klotho deficiency. Taken together, the data presented by Fernandez et al. establish the BECN1-BCL2 complex as target to improve health-span and promote longevity in mammals.
Improving our knowledge on the specific molecular mechanisms regulating autophagy and how this can be modulated to increase the autophagic flux is therefore envisioned to have a big impact in ameliorating healthy ageing. This may indeed lead to the discovery of novel strategies for genetic manipulation of the autophagy process, which, as compared to pharmacological interventions with more pleiotropic effects, clearly provides more elegant and neat ways to specifically modulate a pathway. Nonetheless, extrinsic interventions such as dietary factors or targeted drugs may represent more feasible and malleable ways to promote autophagy. Interestingly, many natural compounds such as flavonoids or polyphenols are known to stimulate autophagy and to have anti-aging effects, although in different experimental settings, and could then represent potential candidates to promote longevity via autophagy induction if a direct causal connection effect will be proved. Most notably, some dietary interventions have been shown to extend lifespan by increasing the autophagic flux (in some cases in an evolutionarily conserved manner) thus representing promising candidates to improve healthy ageing in mammals [(4,5) and references therein] (Table 1). For most of these interventions, however, the exact underlying molecular mechanisms and intracellular targets remain largely undisclosed. Thus, on the one hand it would be interesting to assess whether any of these autophagy stimulators can promote longevity via modulation of the BECN1-BCL2 autophagy regulatory complex; and whether this also has direct repercussion on apoptosis, a form of cell death intrinsically connected with autophagy with important implications for the ageing process as well as for the treatment of age-associated diseases, such as cancer or neurodegenerative disorders. On the other hand, by assessing the effect of BH3 mimetic peptides and small molecules on health- and lifespan extension, we may be able to achieve very specific strategies to promote healthy ageing via autophagy induction.
Table 1
| Intervention | Organism | Reference* |
|---|---|---|
| Dietary restriction | Yeasts, worms, mice | (4,5) |
| Resveratrol | Worms | (4,5) |
| Spermidine | Yeasts, worms, flies | (4,5) |
| Rapamycin | Yeasts, worms, flies | (4,5) |
| Urolithin A | Worms | (4) |
| 2,2’-dipyridyl | Worms | (7) |
| Calcineurin | Worms | (4) |
| Metformin | Flies, mice | (4) |
*And references therein. We apologize for all the original papers we could not cite due to space limitation.
Acknowledgments
Funding: N Ventura’s laboratory is supported by grants from Deutsche Forschungsgemeinschaft (VE 663/3-1, VE 663/6-1, VE 663/8-1) and the Bundesministerium für Bildung und Forschung (HDHL/JPI project MiTyrAge). B Stork’s laboratory is supported by grants from the Deutsche Forschungsgemeinschaft (STO 864/4-1, STO 864/5-1 and GRK 2158), the Research Committee of the Medical Faculty of the Heinrich Heine University Düsseldorf (22/2015), and the Düsseldorf School of Oncology (funded by the Comprehensive Cancer Center Düsseldorf/Deutsche Krebshilfe and the Medical Faculty of the Heinrich-Heine-University Düsseldorf).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Executive Editor-in-Chief Dr. Hualin Sun (Jiangsu Key Laboratory of Neuroregeneration, Nantong University, Nantong, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2018.10.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torgovnick A, Schiavi A, Maglioni S, et al. Healthy aging: what can we learn from Caenorhabditis elegans? Z Gerontol Geriatr 2013;46:623-8. [Crossref] [PubMed]
- Lopez-Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell 2013;153:1194-217. [Crossref] [PubMed]
- Tigges J, Krutmann J, Fritsche E, et al. The Hallmarks of Fibroblast Ageing. Mech Ageing Dev 2014;138:26-44. [Crossref] [PubMed]
- Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 2018;19:579-93. [Crossref] [PubMed]
- Madeo F, Zimmermann A, Maiuri MC, et al. Essential role for autophagy in life span extension. J Clin Invest 2015;125:85-93. [Crossref] [PubMed]
- Markaki M, Palikaras K, Tavernarakis N. Novel Insights Into the Anti-aging Role of Mitophagy. Int Rev Cell Mol Biol 2018;340:169-208. [Crossref] [PubMed]
- Schiavi A, Maglioni S, Palikaras K, et al. Iron-Starvation-Induced Mitophagy Mediates Lifespan Extension upon Mitochondrial Stress in C. elegans. Curr Biol 2015;25:1810-22. [Crossref] [PubMed]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 2008;4:600-6. [Crossref]
- Melendez A, Talloczy Z, Seaman M, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003;301:1387-91. [Crossref] [PubMed]
- Wilhelm T, Byrne J, Medina R, et al. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev 2017;31:1561-72. [Crossref] [PubMed]
- Ejlerskov P, Ashkenazi A, Rubinsztein DC. Genetic enhancement of macroautophagy in vertebrate models of neurodegenerative diseases. Neurobiol Dis 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Longo VD, Antebi A, Bartke A, et al. Interventions to Slow Aging in Humans: Are We Ready? Aging Cell 2015;14:497-510. [Crossref] [PubMed]
- Fernandez AF, Sebti S, Wei Y, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018;558:136-40. [Crossref] [PubMed]
- Hurley JH, Young LN. Mechanisms of Autophagy Initiation. Annu Rev Biochem 2017;86:225-44. [Crossref] [PubMed]
- Levine B, Liu R, Dong X, et al. Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol 2015;25:533-44. [Crossref] [PubMed]
- Mercer TJ, Gubas A, Tooze SA. A molecular perspective of mammalian autophagosome biogenesis. J Biol Che 2018;293:5386-95. [Crossref] [PubMed]
- Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998;72:8586-96. [PubMed]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005;122:927-39. [Crossref] [PubMed]
- Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011;18:571-80. [Crossref] [PubMed]
- Decuypere JP, Parys JB, Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells 2012;1:284-312. [Crossref] [PubMed]
- Gurkar AU, Chu K, Raj L, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun 2013;4:2189. [Crossref] [PubMed]
- Maejima Y, Kyoi S, Zhai P, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013;19:1478-88. [Crossref] [PubMed]
- Zalckvar E, Berissi H, Mizrachy L, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009;10:285-92. [Crossref] [PubMed]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010;3:ra42. [Crossref] [PubMed]
- Rocchi A, Yamamoto S, Ting T, et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer's disease. PLoS Genet 2017;13:e1006962 [Crossref] [PubMed]
Cite this article as: Stork B, Ventura N. Targeting the BECN1-BCL2 autophagy regulatory complex to promote longevity. Biotarget 2018;2:16.


