
The complexity of the ubiquitination code is driven by bacteria
Protein ubiquitination is a post-translational modification dynamically regulated, and is involved in many processes to maintain a balanced functioning of cellular pathways. Since the initial description of the ubiquitin proteasome system (UPS) as a protein destruction mechanism, novel functions have been discovered, including transcriptional regulation, DNA repair, protein-protein interactions (1,2). The complexity in the ubiquitin system has led to the generation of a ubiquitin code, ruled by several and interdependent dynamics that finely regulate the attachment and the detachment of the ubiquitin. The first step of this process is the coupling of a ubiquitin molecule on a Lysine (Lys) residue localized on the substrate; then, the isopeptide-linked ubiquitin chains can be generated, exploiting all the seven Lys residues of the ubiquitin that can be ubiquitinated (3). The kinds of linkages in the ubiquitin chain determine the destiny of the marked protein. Three enzymes perform the ubiquitin reaction: the ubiquitin-activating enzyme E1, the ubiquitin-activating enzyme E2, and the ubiquitin ligase E3, that exhibits substrate specificity. In eukaryotic cells, there are two major types of E3 ubiquitin ligases: homologous to the E6-AP carboxyl terminus (HECT)-type that form a thioester intermediate with ubiquitin before transferring it to the substrate, and really interesting new genes (RING)/U-box types, which work as scaffolds promoting the interaction between E2s and the targeted substrates (4). One of the most important role of the ubiquitin system is that of a host cell defense mechanism against viral and bacterial infections, acting by recruiting inflammatory cells and stimulating antigen presentation (5). Furthermore, ubiquitin marks bacteria for killing them by both the proteasome and autophagy-mediated degradation systems (6). In order to counteract microbicidal programs, bacteria have evolved survival strategies against host ubiquitin system, by manipulating component of the host protein modification pathways during infection (7,8). Recently, bacterial ubiquitin ligase-like effectors (proteins that mimic the function of the host E3 ubiquitin ligase) are found to be crucial for the bacterial life cycle. Among gram-negative bacteria, Legionella pneumophila has the largest group of effector proteins (more than 300) and at least 10 proteins are involved in ubiquitin manipulation (9,10). The SidE family of Legionella pneumophila effectors is a peculiar group of ubiquitin-modifying enzymes. This family includes four large proteins (SidE, SdeA, SdeB, and SdeC) that are essential for its replication in host cells (11,12). SidE proteins are formed by four domains: a deubiquitinase (DUB) domain, a phosphodiesterase (PDE) domain, a mono-ADP-ribosyltransferase (mART) domain, and a coiled-coil (CC) domain (Figure 1A). Intriguingly, while all known bacterial E3 ligases need ATP-dependent E1 and E2 activity, SdeA regulates the attachment of ubiquitin autonomously (13,14). In more details, the mART domain uses the cofactor nicotinamide adenine dinucleotide (NAD) to form a phospho-ribosylated ubiquitin (Pr-Ub) that is then attached to the substrates on their serine residues through a phosphor-ribosyl linkage. More importantly, intracellular mono-ADP-ribosylation leads to an impairment of central ubiquitin-dependent pathways, such as cell proliferation, TNF signaling and mitophagy (14). A recent study by Kalayil et al. set out to describe the complexity of the molecular architecture of the SdeA-drived catalytic platform (15). First, they crystallized the minimal fragment of SdeA (residues 213-907) that is necessary for the ubiquitination of its substrates (Figure 1A). This fragment of the protein comprises both the PDE and mART domains of SdeA, this suggesting the presence of two catalytic sites for substrate modification. Detailed analysis of the structure reveals that the mART catalytic site is formed by a α-helical lobe (AHL) and the mART core, respectively. Surprisingly, at variance with the structure of other bacterial ADP-ribosylating enzymes, the AHL has no physical proximity to the mART core; this prompted Kalayil et al. to investigate and demonstrate the existence of a transient conformation for NAD+ binding and processing (Figure 1B).
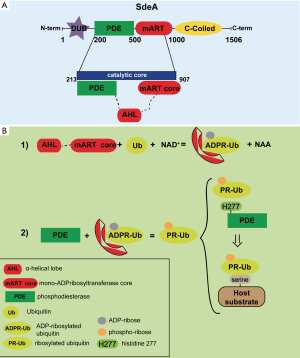
About the catalytic site in the PDE, since the pocket is enriched with conserved histidines, the authors seeked for the presence of a transient intermediate implicating covalent binding of phosphate to a catalytic histidine residue. In more details, they identified a two-step phosphoribosyl-ubiquitin transfer reaction that involves the histidine H277 of SdeA. First, H277 is attached by phosphoribose to ubiquitin through a phosphoramidate bond forming a transient SdeA H277-PR-Ub intermediate; next, ubiquitin is transferred to serine residues of the substrate via a phosphoribose linker (Figure 1B) (14). The existence of a transient intermediate is sustained by biochemical data reported in an accompanying paper (16).
After that, Kalayil and co-authors aimed at determining the ubiquitination sites within the SdeA substrate RTN4B. By mass spectrometry approaches, they identified two ubiquitination sites on RTN4B, containing two serine residues. By means of sequence alignment of RTN4B peptides, they found that SdeA promotes the ubiquitination of hydrophobic residues surrounding the target serine residues and they are usually within disordered regions. This opens the possibility that SdeA could target disordered serine residues in many host substrates. Based on these findings, and by diving into the understanding of what mechanism (phosphoribosylation of ubiquitin or substrate ubiquitination) was physiological fundamental, the authors demonstrated by mutagenesis approaches the importance of the specific ubiquitination of a substrate for bacterial pathogenicity, rather than a ubiquitin modification.
In summary, we can consider SdeA as a peculiar bacterial virulence protein that is able to foster phosphoribosylation of ubiquitin. The dissection of the SdeA activity has impact not only on the understanding of basic mechanism of this uncommon post-translational modification, but moreover provides promising therapeutic approaches for targeting bacteria at distinct stages of infection. However, since many Legionella effector proteins have evolutionarily eukaryotic origins (17), it could be exciting to discover an equivalent machinery in eukaryotes, adding more complexity and tangle to the ubiquitin code.
Acknowledgments
Funding: F. Cecconi’s laboratory is supported by grants from the Danish Cancer Society (KBVU R72-A4408, R146-A9364), the Novo Nordisk Foundation (7559, 22544), the European Union (Horizon 2020 MEL-PLEX, 642295) the Lundbeckfonden (R233-2016-3360), the LEO Foundation (LF17024), the Bjarne Saxhof Foundation and the AIRC Investigator Grant (IG2016–18906). Further, F. Cecconi’s lab in Copenhagen is part of the Center of Excellence for Autophagy, Recycling and Disease (CARD funded by the Danish National Research Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Executive Editor-in-Chief Dr. Hualin Sun (Jiangsu Key Laboratory of Neuroregeneration, Nantong University, Nantong, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2018.08.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol 2015;16:322-4. [Crossref] [PubMed]
- Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol 2016;18:579-86. [Crossref] [PubMed]
- Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci 2016;129:875-80. [Crossref] [PubMed]
- Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 2017;86:129-57. [Crossref] [PubMed]
- Li J, Chai QY, Liu CH. The ubiquitin system: a critical regulator of innate immunity and pathogen-host interactions. Cell Mol Immunol 2016;13:560-76. [Crossref] [PubMed]
- Yuk JM, Yoshimori T, Jo EK. Autophagy and bacterial infectious diseases. Exp Mol Med 2012;44:99-108. [Crossref] [PubMed]
- Zhou Y, Zhu Y. Diversity of bacterial manipulation of the host ubiquitin pathways. Cell Microbiol 2015;17:26-34. [Crossref] [PubMed]
- Maculins T, Fiskin E, Bhogaraju S, et al. Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res 2016;26:499-510. [Crossref] [PubMed]
- Qiu J, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 2017;15:591-605. [Crossref] [PubMed]
- Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 2010;26:261-83. [Crossref] [PubMed]
- Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 2005;56:90-103. [Crossref] [PubMed]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 2004;101:841-6. [Crossref] [PubMed]
- Qiu J, Sheedlo MJ, Yu K, et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 2016;533:120-4. [Crossref] [PubMed]
- Bhogaraju S, Kalayil S, Liu Y, et al. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 2016;167:1636-49.e13. [Crossref] [PubMed]
- Kalayil S, Bhogaraju S, Bonn F, et al. Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 2018;557:734-8. [Crossref] [PubMed]
- Akturk A, Wasilko DJ, Wu X, et al. Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector. Nature 2018;557:729-33. [Crossref] [PubMed]
- Burstein D, Amaro F, Zusman T, et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 2016;48:167-75. [Crossref] [PubMed]
Cite this article as: Nazio F, Cecconi F. The complexity of the ubiquitination code is driven by bacteria. Biotarget 2018;2:15.


