
NKX3.2 plays a key role in regulating HIF1α-directed angiogenesis in chondrocytes
The cellular fraction of cartilage is mainly composed of one single cell type, the chondrocyte, which secretes, shapes and maintains the cartilaginous matrix that functions, in the case of epiphyseal cartilage, as the template for bone elongation. The biomechanical properties of cartilage are dependent mainly on the composition, as well as the macromolecular integrity of its matrix contributing to the functional and phenotypic differences between cartilage subtypes (1). For example, articular cartilage (AC) is a highly resilient tissue that allows low-friction joint articulation. In contrast, growth plate (GP) cartilage is populated by highly proliferative chondrocytes that undergo hypertrophic differentiation in an angiogenesis-promoting spatiotemporal manner, serving as a mold for longitudinal bone growth. As such, an in sharp contrast to AC, GP cartilage is a transient tissue and is replaced by bone in a process known as endochondral ossification (2).
Hypoxia is a major stimulus of chondrogenesis as well as angiogenesis, by stabilizing hypoxia inducible factor 1α (HIF1α) (3). HIF1α acts as a transcriptional activator of SOX9, and of vascular endothelial growth factor (VEGF) expression, promoting angiogenesis (4). Cartilage is hypoxic in nature and angiogenesis-dependent reoxygenation of GP cartilage is a major trigger for its hypertrophic differentiation. While HIF1α-induced expression of VEGF and subsequent angiogenesis is a normal process in GP hypertrophic differentiation, AC, while hypoxic in nature, appears to be resistant to hypoxia-stimulated angiogenesis and subsequent hypertrophic differentiation. The specific resistance of AC to hypertrophic differentiation has been ascribed to various soluble factors, such as parathyroid hormone related protein (PTHrP) (5), as well as the WNT antagonists DKK1 and FRZB that are highly expressed in AC, but not in the GP (6). We have previously found that expression of DKK1 and FRZB is highly upregulated in chondrogenically differentiating mesenchymal stem cells in hypoxic conditions (7,8). Another important regulator for the prevention of chondrocyte hypertrophy is NKX3.2. NKX3.2 is a homeobox transcriptional regulator (9), and a downstream transcriptional effector of SHH (10), PTHrP (11), and bone morphogenic protein (BMP)-7 (12), and is expressed as a target of SOX9 activity (13) (Figure 1A). It inhibits chondrocyte hypertrophy by repressing RUNX2 activity (14) and in concert with this notion genetic deletion of NKX3.2 or overexpression (15) induce severe developmental skeletal abnormalities which are ascribed to altered chondrocyte maturation. The inactivating mutation in the NKX3.2 gene causing the human skeletal condition SMMD further underscores its critical role in chondrocyte development (16). Therefore, NKX3.2 is considered an important mediator in preventing the switch to chondrocyte hypertrophy (17). While the above molecular mediators provide avenues to prevent chondrocyte hypertrophy of AC, these mechanisms do not explain why VEGF-induced angiogenesis is not present in the hypoxic environment of cartilage tissue.
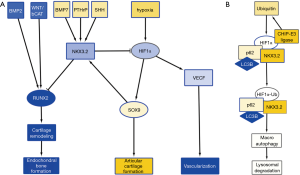
A recent paper in Cellular Signaling by Suhjean Im and Dae-Won Kim (18) describes a mechanism by which NKX3.2 may prevent angiogenic activity by inducing target-specific HIF1α degradation in chondrocytes in the hypoxic cartilage environment. In a series of experiments, they uncover a novel NKX3.2-directed pathway that results in an oxygen-concentration independent and proteasome-independent degradation of HIF1α.
In an initial experiment, the authors tested the hypothesis that NKX3.2 could modulate HIF1α protein expression, thereby reducing a major vascularization trigger, typically found in hypoxic tissues. Overexpression of NKX3.2 in cells cultured in hypoxia, reduced the expression of HIF1α. Inversely, knockdown of NKX3.2 resulted in elevated levels of HIF1α.
Next, the authors investigated the mechanism of HIF1α degradation. It has been described that HIF1α is targeted for destruction by an E3 ubiquitin ligase, containing the von Hippel-Lindau tumor suppressor protein (pVHL) (19). In this pVHL-mediated proteasomal degradation pathway, proline hydroxylation is essential. However, Im and Kim found that this pathway was not involved in the NKX3.2-mediated HIF1α degradation. Moreover, they found that NKX3.2-induced HIF1α degradation was proteasome independent. In a subsequent experiment they showed that in chondrocytes, both in normoxia and hypoxia, NKX3.2 induced HIF1α degradation via a lysosomal pathway, specifically via macroautophagy.
Autophagy is a degradative pathway that occurs in all eukaryotic cells. Its main function is to recycle cytoplasm to generate macromolecular building blocks and energy under stress conditions, and to remove superfluous and damaged organelles to enable adaptation to changing nutrient conditions, as well as to maintain cellular homeostasis (20). Macroautophagy is a specific type of autophagy in which the substrates are sequestered within cytosolic double membranes that are called autophagosomes (20). The autophagosome fuses with lysosomes and the contents are degraded and recycled.
Macroautophagy can be a selective process, regulated by specific cargo adaptor proteins such as p62/SQSTM1, Nbr1 and BNIP3, that then bind to ATG8 homologs, such as: LC3A and LC3B, to induce autophagy [reviewed in (21), and articles in issue 16 of Nature Cell Biology]. Im and Kim investigated whether NKX3.2 modulates the interaction between HIF1α and these adaptor proteins. They found that p62/SQSTM1 and LC3B are involved in the autophagy of HIF1α (Figure 1B). In addition, they found that ubiquitination of HIF1α by CHIP-E3 ligase, was elevated by NKX3.2 expression. This ubiquitination however, did not lead to proteasomal degradation, but led to association of HIF1α with p62/SQSTM1 and its subsequent degradation by macroautophagy. CHIP had been previously identified as the E3 ligase involved in HIF1α degradation in normoxic conditions (22,23). This makes us wonder why ubiquitination of HIF1α by CHIP does not lead to proteasomal degradation but rather macroautophagy in hypoxic conditions when NKX3.2 is involved?
The findings in this manuscript indicate that NKX3.2-mediated regulation of HIF1α stability may play a role in the homeostasis of AC, by controlling angiogenic signals and keeping vascularization in pace, thereby contributing in the maintenance of a permanent AC phenotype.
What are the potential implications of this finding?
As described above, many papers, including our own work (12,13,15,17), have shown that NKX3.2 expression is associated with reduced chondrocyte hypertrophy, and thus maintenance of a healthy AC phenotype. Taking the mechanism into consideration described by Im and Kim, this would imply that under healthy cartilage conditions where NKX3.2 expression is relatively high, HIF1α would be more effectively targeted for degradation. However, this seems in sharp contradiction with the well-described notion that increased HIF1α levels are beneficial for cartilage homeostasis, not in the least place by regulating expression of the cartilage master transcription factor SOX9 (24) (Figure 1A). The question arises what the order of events is in regulating the expression of HIF1α, SOX9, NKX3.2, RUNX2, etc. One could postulate that it is a matter of gradients as depicted in Figure 2, where an intricate balance between multiple factors regulates the exact spatiotemporal differentiation of the different zones of the cartilage in the GP. As can be seen in Figure 2, HIF1α expression does not necessarily correlate to the regions of hypoxia. In the hypertrophic zone (HZ), close to the primary spongiosum, HIF1α expression is higher than can be explained by the absence of oxygen (Figure 2). It important to note that most studies on HIF1α expression in the developing limb, are based on the use of markers for hypoxia, such as EF-5, rather than antibody-based staining of HIF1α (25). The actual HIF1α expression pattern in the GP as depicted in Figure 2 is therefore AN interpretation of the various studies combining staining of hypoxia and HIF1α in sections of the GP.
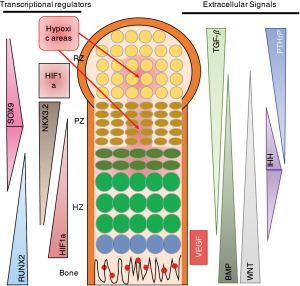
The expression pattern of NKX3.2 would explain the absence of vascularization of the hypoxic areas in the resting zone (RZ) and proliferative zone (PZ). However, the absence of HIF1α in these areas would also suggest a reduction of the amount of SOX9 expression, as SOX9 is a target gene of HIF1α (24). This however, is not the case. One could speculate that this is due to the fact that SOX9 is able to regulate its own expression via a positive feedback loop with SOX 5/6, which has been shown to be sufficient for cartilage development (28). This is substantiated by the finding that SOX9 expression was independent of the oxygen concentration (29).
Another possible explanation of the findings is that timing is critical in the regulation of cartilage and endochondral ossification. It is possible that HIF1α activates SOX9 in the onset of limb formation, when HIF1α is expressed homogeneously throughout the limb bud (11). SOX9 then activates SOX5/6 expression and can, via a positive feedback loop, ensure its own transcription. During later stages, when the fetal primary GP with the various cartilage zones is formed, there are more distinct expression patterns of the various factors involved in regulation of proliferation, differentiation and hypertrophy (Figure 2). In these different zones of the GP, an intricate network of signals regulates the various stages of endochondral ossification? (26). Interestingly, NKX3.2 starts to be expressed in the developing limb at stage E10.5 and at later developmental stages is mainly expressed in the PZ and decreases towards the HZ (Figure 2) (30). As most studies investigating the role of NKX3.2 in cartilage have been performed in the prenatal and early postnatal stages, it is hard to speculate what the role of NKX3.2 is in other endochondral ossification-related processes like the secondary ossification center and in fracture healing.
We conclude that the NKX3.2-guided mechanism of HIF1α degradation as described by Im and Kim (18), is an interesting finding with respect to protecting cartilage to angiogenic signals, thereby maintaining cartilage homeostasis. However, this finding leads to many unanswered questions considering the role of hypoxia and HIF1α expression as a mechanism of controlling chondrocyte hypertrophy by regulating expression and activity of the pro-cartilage factor SOX9.
Acknowledgments
Funding: Marcel Karperien and Janine Post acknowledge center of excellence funding from the Dutch Arthritis Society (grant #LLP-25).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Editor-in-Chief Maorong Jiang, MD (Laboratory Animal Center, Nantong University, Nantong, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2018.07.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ahmed TA, Hincke MT. Strategies for Articular Cartilage Lesion Repair and Functional Restoration. Tissue Eng Part B Rev 2010;16:305-29. [Crossref] [PubMed]
- Mackie EJ, Ahmed YA, Tatarczuch L, et al. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol 2008;40:46-62. [Crossref] [PubMed]
- Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev 2001;11:293-9. [Crossref] [PubMed]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721-32. [Crossref] [PubMed]
- Karp SJ, Schipani E, St-Jacques B, et al. Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 2000;127:543-8. [PubMed]
- Leijten JC, Emons J, Sticht C, et al. Gremlin 1, frizzled-related protein, and Dkk-1 are key regulators of human articular cartilage homeostasis. Arthritis Rheum 2012;64:3302-12. [Crossref] [PubMed]
- Leijten J, Georgi N, Moreira Teixeira L, et al. Metabolic programming of mesenchymal stromal cells by oxygen tension directs chondrogenic cell fate. Proc Natl Acad Sci U S A 2014;111:13954-9. [Crossref] [PubMed]
- Leijten JC, Moreira Teixeira LS, Landman EB, et al. Hypoxia inhibits hypertrophic differentiation and endochondral ossification in explanted tibiae. PLoS One 2012;7:e49896 [Crossref] [PubMed]
- Chatterjee S, Kraus P, Sivakamasundari V, et al. Genome wide binding (ChIP-Seq) of murine Bapx1 and Sox9 proteins in vivo and in vitro. Genom Data 2016;10:51-3. [Crossref] [PubMed]
- Daoud G, Kempf H, Kumar D, et al. BMP-mediated induction of GATA4/5/6 blocks somitic responsiveness to SHH. Development 2014;141:3978-87. [Crossref] [PubMed]
- Provot S, Kempf H, Murtaugh LC, et al. Nkx3.2/Bapx1 acts as a negative regulator of chondrocyte maturation. Development 2006;133:651-62. [Crossref] [PubMed]
- Caron MM, Emans PJ, Cremers A, et al. Hypertrophic differentiation during chondrogenic differentiation of progenitor cells is stimulated by BMP-2 but suppressed by BMP-7. Osteoarthritis Cartilage 2013;21:604-13. [Crossref] [PubMed]
- Yamashita S, Andoh M, Ueno-Kudoh H, et al. Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp Cell Res 2009;315:2231-40. [Crossref] [PubMed]
- Lengner CJ, Hassan MQ, Serra RW, et al. Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. J Biol Chem 2005;280:15872-9. [Crossref] [PubMed]
- Jeong DU, Choi JY, Kim DW. Cartilage-Specific and Cre-Dependent Nkx3.2 Overexpression In Vivo Causes Skeletal Dwarfism by Delaying Cartilage Hypertrophy. J Cell Physiol 2017;232:78-90. [Crossref] [PubMed]
- Hellemans J, Simon M, Dheedene A, et al. Homozygous inactivating mutations in the NKX3-2 gene result in spondylo-megaepiphyseal-metaphyseal dysplasia. Am J Hum Genet 2009;85:916-22. [Crossref] [PubMed]
- Caron MM, Emans PJ, Surtel DA, et al. BAPX-1/NKX-3.2 acts as a chondrocyte hypertrophy molecular switch in osteoarthritis. Arthritis Rheumatol 2015;67:2944-56. [Crossref] [PubMed]
- Im S, Kim DW. Nkx3.2 induces oxygen concentration-independent and lysosome-dependent degradation of HIF-1α to modulate hypoxic responses in chondrocytes. Cell Signal 2017;36:127-38. [Crossref] [PubMed]
- Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292:464-8. [Crossref] [PubMed]
- Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Research 2014;24:24-41. [Crossref] [PubMed]
- Johansen T, Lamark T. Selective autophagy goes exclusive. Nature Cell Biology 2014;16:395. [Crossref] [PubMed]
- Ferreira JV, Fôfo H, Bejarano E, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy 2013;9:1349-66. [Crossref] [PubMed]
- Luo W, Zhong J, Chang R, et al. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1alpha but Not HIF-2alpha. J Biol Chem 2010;285:3651-63. [Crossref] [PubMed]
- Amarilio R, Viukov SV, Sharir A, et al. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development 2007;134:3917-28. [Crossref] [PubMed]
- Schipani E, Mangiavini L, Merceron C. HIF-1alpha and growth plate development: what we really know. Bonekey Rep 2015;4:730. [Crossref] [PubMed]
- Kerkhofs J, Roberts SJ, Luyten FP, et al. Relating the chondrocyte gene network to growth plate morphology: from genes to phenotype. PLoS One 2012;7:e34729 [Crossref] [PubMed]
- Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 2016;4:16009. [Crossref] [PubMed]
- Ikeda T, Kamekura S, Mabuchi A, et al. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum 2004;50:3561-73. [Crossref] [PubMed]
- Kawato Y, Hirao M, Ebina K, et al. Nkx3.2-induced suppression of Runx2 is a crucial mediator of hypoxia-dependent maintenance of chondrocyte phenotypes. Biochem Biophys Res Commun 2011;416:205-10. [Crossref] [PubMed]
- Rainbow RS, Won HK, Zeng L. The role of Nkx3.2 in chondrogenesis. Front Biol (Beijing) 2014;9:376-81. [Crossref] [PubMed]
Cite this article as: Welting TJM, Karperien M, Post JN. NKX3.2 plays a key role in regulating HIF1α-directed angiogenesis in chondrocytes. Biotarget 2018;2:11.


