
Regulation of SCF E3 ligase activity by Cand1
Ubiquitination, which targets proteins for degradation or modifies their activity, is central to cellular function. Proteins controlling ubiquitination have attracted a lot of attention as potential therapeutic targets for a variety of diseases. In particular, E3 ligases are responsible for attachment of ubiquitin (Ub) to target proteins, and thus, dictate specificity of ubiquitination and could be targets for therapeutic intervention. Among more than 600 E3 ligases in the human genome, Cullin Ring Ligases (CRLs) constitute the largest family with approximately 200 members (1). CRLs share a common architecture, in which a Cullin subunit is responsible for tethering a substrate receptor, and a RING protein recruits E2-Ub. The family of human Skp1-F-box-Cul1 ligases (SCFs), the best characterized CRL family, contains 69 F-box proteins that are responsible for substrate binding and are attached to a cullin subunit (Cul1) through the adaptor Skp1 (2).
The functions of cullin scaffolds extend far beyond simply bringing substrate receptors and RING-E2-Ub together. All cullin scaffolds are modified by attachment of a Ub-like modifier, Nedd8, which serves to re-orient RING protein and increase the efficiency of ubiquitination (3,4). Importantly, the presence of a substrate protein undergoing ubiquitination inhibits Nedd8 removal by Cop9-signalosome (5,6), providing a positive feedback loop for the ubiquitination reaction. An additional layer of CRL regulation is provided by the Cand1 protein. In the absence of Nedd8 modification, Cand1 wraps around the cullin subunit and blocks the recruitment of substrate receptor proteins (7,8).
Although Cand1 appears to act as inhibitor that blocks assembly of CRL ligases, it was observed in multiple studies that deletion of Cand1 in cells actually decreases CRL activity (9,10). This conundrum was explained by several studies focused on human (11) and yeast (12,13) SCF E3 ligases. These studies demonstrated that Cand1 functions as an exchange factor, by freeing Cul1 from unproductive SCF complexes (Figure 1). Neddylation of Cul1 plays a crucial role in this process, as upon substrate removal, Cul1 is deneddylated and Cand1 is able to bind. The binding of Cand1 in turn removes the Skp1-F-box component and liberates Cul1 for binding to a different Skp1-F-box complex. Importantly, this mechanism favors assembly of SCF complexes for which substrates are available, and this has been experimentally confirmed (14).

A recent study by Liu et al. (15) set out to describe the complexity of SCF regulation by neddylation and Cand1 binding with a mathematical model that takes into account all known aspects of SCF regulation, protein concentrations and kinetic parameters of protein interactions, some of which were derived in the study. Highlighting the complexity of SCF regulation, the authors serendipitously discovered that the binding of Cand1 actually increases association of Cul1 with Dcn1, the E3 ligase responsible for Nedd8 addition. The authors suggested that this primes Cul1 for neddylation immediately upon Cand1 removal, and indeed, Cand1-Cul1 complex in the presence of Skp1-F-box complexes is neddylated more efficiently then Cul1 alone. This positive cooperativity between Cand1 and Dcn1 was also included in the model.
The developed mathematical model accurately predicted several experimental observations that would be difficult to anticipate otherwise. For example, the model correctly predicted that the degradation defect of β-TrCP substrate in Cand1 deleted cells is fully rescued by re-expression of Cand1 at only 13% of wild-type levels. Another counter-intuitive prediction of the model was that the defect in the degradation of β-TrCP substrate (IκBα) upon Cand1 deletion could be rescued by Cul1 overexpression, but not by β-Trcp overexpression. This prediction was experimentally confirmed and further highlighted Cul1 as a limiting factor for SCF activity and the importance of Cand1 exchange activity for liberating Cul1 from unproductive complexes.
A striking prediction of the model is that if Cul1 binds an F-box protein unoccupied by substrate, it is able to exchange it for another F-box protein with an average time of 87 seconds (Figure 2). The authors suggest that this allows Cul1 to rapidly sample through the whole pool of cellular F-box proteins and, given a ratio of Skp1 to Cul1 of 4:1 (14), an F-box protein should gain access to Cul1 approximately every four minutes. Once Cul1 is engaged with an F-box protein with a bound substrate, it is removed from the rapidly cycling pool of Cul1 and the SCF complex persists until the substrate is degraded. This numerical description of Cul1 function re-enforces the “on demand” concept introduced in earlier studies, which states that Cul1 distribution is biased towards those F-box proteins that are needed at the moment.
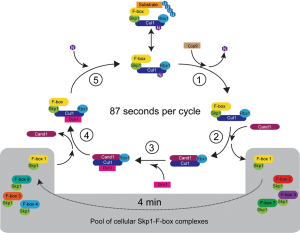
Another important prediction of the model is that cells without Cand1 function are particularly sensitive to variations in the levels of F-box proteins. This was experimentally verified by showing that cells lacking Cand1 (and Cand2 that can partially compensate for Cand1 function) exhibited significant growth defects and abnormal morphology upon over-expression of F-box proteins. It would be interesting to further explore this observation by testing whether cancer cells that are known to over-express F-box proteins (16,17) are particularly sensitive to Cand1 deletion, which may identify Cand1 as a potential therapeutic target. While this study relied on deletion of the Cand1 gene, it would also be worthwhile to use the developed model coupled with the experimental validation to explore the effects of inhibiting Cand1 binding to Cul1. In particular, it would be informative to compare the effects of targeting different protein interaction surfaces to abrogate the interaction between Cand1 and Cul1. While it is difficult to develop chemical compounds to block protein interactions, research by our group developed the use of Ub variants (UbVs) to target proteins of the ubiquitin proteasome system (18), and this approach could be extended to investigate the effects of blocking Cand1 interactions.
In summary, the mathematical model developed by Liu et al. (15)was able to accurately predict response of SCF activity to different perturbations. The model can serve as a powerful tool to investigate different sensitivities of SCF activity, which may be useful for therapeutic applications. For example, the small-molecule MLN4924 inhibits neddylation and is currently being investigated in clinical trials for treatment of various cancers (19). Researches may ask what new sensitivity is introduced into the system upon treatment with MLN9294 and could potentially uncover new avenues for therapeutic intervention.
Acknowledgments
Funding: This work was supported by Canadian Institutes of Health Research (CIHR) grants.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Biotarget. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2018.05.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bhowmick P, Pancsa R, Guharoy M, et al. Functional diversity and structural disorder in the human ubiquitination pathway. PLoS One 2013;8:e65443 [Crossref] [PubMed]
- Jin J, Cardozo T, Lovering RC, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 2004;18:2573-80. [Crossref] [PubMed]
- Duda DM, Borg LA, Scott DC, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 2008;134:995-1006. [Crossref] [PubMed]
- Jones J, Wu K, Yang Y, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res 2008;7:1274-87. [Crossref] [PubMed]
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci U S A 2006;103:11515-20. [Crossref] [PubMed]
- Cavadini S, Fischer ES, Bunker RD, et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 2016;531:598-603. [Crossref] [PubMed]
- Zheng J, Yang X, Harrell JM, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 2002;10:1519-26. [Crossref] [PubMed]
- Goldenberg SJ, Cascio TC, Shumway SD, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 2004;119:517-28. [Crossref] [PubMed]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol 2006;26:1235-44. [Crossref] [PubMed]
- Cheng Y, Dai X, Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol 2004;135:1020-6. [Crossref] [PubMed]
- Pierce NW, Lee JE, Liu X, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 2013;153:206-15. [Crossref] [PubMed]
- Zemla A, Thomas Y, Kedziora S, et al. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun 2013;4:1641. [Crossref] [PubMed]
- Wu S, Zhu W, Nhan T, et al. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun 2013;4:1642. [Crossref] [PubMed]
- Reitsma JM, Liu X, Reichermeier KM, et al. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell 2017;171:1326-39.e14. [Crossref] [PubMed]
- Liu X, Reitsma JM, Mamrosh JL, et al. Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression. Mol Cell 2018;69:773-86 e6.
- Vaidyanathan S, Cato K, Tang L, et al. In vivo overexpression of Emi1 promotes chromosome instability and tumorigenesis. Oncogene 2016;35:5446-55. [Crossref] [PubMed]
- Zhang W, Cao L, Sun Z, et al. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle 2016;15:1344-51. [Crossref] [PubMed]
- Ernst A, Avvakumov G, Tong J, et al. A strategy for modulation of enzymes in the ubiquitin system. Science 2013;339:590-5. [Crossref] [PubMed]
- Nawrocki ST, Griffin P, Kelly KR, et al. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs 2012;21:1563-73. [Crossref] [PubMed]
Cite this article as: Gorelik M, Sidhu SS. Regulation of SCF E3 ligase activity by Cand1. Biotarget 2018;2:10.


