
Long noncoding RNAs regulate malignant phenotypes in colorectal cancer
Introduction
Because colorectal cancer (CRC) is the fourth most frequent type of malignancy and because the incidence of CRC is increasing worldwide, this disease is a serious threat to human health (1,2). Researchers have determined the mechanisms of CRC, reducing the mortality from CRC in the past several decades. However, the prognosis of this malignancy remains poor, and the development of new biomarkers and therapeutic strategies is challenging.
Recent advances in sequencing technologies have revealed that protein-coding genes only occupy 3% of the human genome, suggesting that noncoding RNAs (ncRNAs) have a crucial role in process between DNAs and proteins (3,4). Long noncoding RNAs (LncRNAs), which have different biological functions and implications, are a category of ncRNAs that are over 200 nucleotides (5). Some lncRNAs are evolutionarily conserved, despite having limited sequence similarity. Syntenic genomic sites frequently encode evolutionarily conserved, functional lncRNAs (6,7).
Several lncRNAs are uniquely expressed and exquisitely regulated in cancer cells (8). Although high-throughput studies have provided instructions on how mutations in protein-coding RNAs genetically affect translation, the biology of ncRNAs, including lncRNAs, has been unknown (9). However, accumulated knowledge on lncRNAs and emerging technologies has enabled researchers to annotate cancer-related lncRNAs and their pathways. Furthermore, Batista et al. reported the importance of the localization of lncRNAs, which influences cell fates during development, and their dysregulation causes human disorders (10). The regulation of lncRNAs at the chromatin level is a well-reported mechanism that affects intra-chromosomal genes and accordingly targets gene transcription in cis or trans (11,12). LncRNAs have been reported as cis enhancers that regulate transcription through chromatin remodeling, methylation, and transcriptional factor trapping (13-15). For example, the lncRNA X-inactive specific transcript (XIST), which is the master regulator of X chromosome inactivation, accumulates in cis on the X chromosome (16). Chu et al. reported that XIST interacts with 81 chromatin modification and RNA remodeling proteins, inducing the transition from pluripotency to differentiation (17).
Further, LncRNAs modulate RNA metabolism by controlling mRNA stability, splicing, and transcription. MALAT-1, also known as NEAT2 and highly conserved in mammals, has been reported to interact with pre-mRNA in the proximal region of the chromatin of a target gene. Through this interaction, MALAT1 regulates gene expression through splicing (18). The lncRNA Staufen-1 (STAU-1) also binds to the 3’UTR region of the target gene, collapsing mRNA (19). This interaction between STAU-1 and mRNA is regulated by terminal differentiation-induced noncoding RNA (TINCR), promoting epidermal differentiation, and is suppressed in poorly differentiated squamous cell carcinoma (20).
lncRNAs interact with protein, inducing changes in protein–protein interactions, protein localization, and protein function—this interaction is thought to be a crucial role of lncRNAs (21). As described above, XIST binds to several proteins, inducing X chromatin inactivation (17). Recent RNA chromatography techniques have enabled researchers to detect this interaction genetically and biochemically. By RNA immunoprecipitation, lncRNA pull-down, and RNA-seq, the interaction between lncRNAs and proteins has been studied extensively (22,23).
Thus, lncRNAs have several functions and affect many pathways in human cells. Here, we focus on lncRNAs and their role in human CRC, as these RNAs are thought to be essential targets for next-generation diagnosis and therapy in CRC in the ncRNA field.
LncRNA regulates cancer phenotypes
Currently, cancer is believed to be regulated by intrinsic cellular dysfunction and intercellular connections with the cancer microenvironment. Based on this concept, cancer phenotypes are divided to six categories during the development of human cancer (24)—proliferation, growth suppressor, motility, immortality, and viability—that affect the growth and metastasis of cancers. In this review, we focus on the function of lncRNAs in each cancer hallmark, describing the related cancer pathways in CRC (Figure 1).
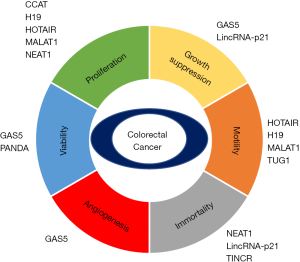
Colon cancer-associated transcript (CCAT) family
Current studies have shown an association of multiple genomic variants, including lncRNAs, in chromosome 8q24, which is specific for c-MYC (25). CCAT1 is a recently discovered lncRNA on chromosome 8q24 that is 2,628 nucleotides and has a short isoform, CCAT1-S, and a long isoform, CCAT1-L. Several studies have revealed the function and mechanisms of CCAT1 in CRC. Kim et al. reported that CCAT1-S, known as Carlo-5, is significantly associated with the cancer-associated variant rs6983267 in the MYC enhancer region in CRC. They also found that the promoter region of CCAT1-S interacts physically with the MYC enhancer region, regulating CCAT1-S expression (26). Recently, CCAT1 was reported to be a target of the bromodomain and extraterminal (BET) protein and marks the colon cMYC super-enhancer. This result indicates that CCAT1 might be a clinical biomarker for detecting CRC patients who respond to a BET inhibitor as chemotherapy (27). Furthermore, the other isoform, CCAT1-L, which is transcribed from a super-enhancer region of MYC, modulates chromatin looping between the MYC promoter and its enhancers by interacting with CCCTC-binding factor (CTCF) (28).
Ling et al. identified CCAT2 and demonstrated that CCAT2 regulates MYC, miR-17-5p and miR-20a through the regulation of transcription factor 7-like 2. They also revealed that CCAT2 is a downstream target gene of WNT, which means that these genes form a feedback loop, and that SNP status influences CCAT2 expression (29). Moreover, Redis et al. showed that CCAT2 regulates cancer metabolism in an allele-specific manner by binding cleavage factor I and affects carcinogenesis in CRC (30).
As biomarkers of CRC, high expression of CCAT1 and CCAT2 has been revealed to be significantly associated with poor overall survival and disease-free survival in 300 CRC samples (31). Thus, CCAT1 and CCAT2 regulate several oncogenic genes and pathways, indicating their essential role in CRC.
H19
The lncRNA H19 was discovered as a parentally imprinted gene and is located on human chromosome 11p15.5 (32,33). H19 is abundantly expressed during embryonic development and is nearly completely repressed after birth (34). Since its discovery, several studies have shown that H19 has an essential role in the progression of multiple types of cancers as an oncogene and tumor suppressor (35-38).
H19 is also known as the precursor gene of miR-675 (39). Tsang et al. revealed that H19 and miR-675 are highly expressed in CRC tissue compared with normal colon tissue. Furthermore, they found that the tumor suppressor retinoblastoma (RB) was a direct target of miR-675, indicating that H19-derived miRNA-675 regulates cancer development in CRC through downregulation of RB (40). Regarding the H19-miR-675 axis, Chen et al. reported that overexpression of H19 increases the resistance to vitamin D treatment by targeting vitamin D receptor through miR-675-5p in CRC (41). H19 recruits the cell cycle-related gene eukaryotic translation initiation factor 4A3 (eIF4A3) and promotes proliferation of CRC cells, based on bioinformatics methods. This study also showed that higher expression of H19 is related to tumor differentiation and advanced TNM stage in CRC patients (42). We reported that H19 is the most significant lncRNA among several cancer-related lncRNAs in The Cancer Genome Atlas CRC dataset. Furthermore, using two other physical CRC datasets, we confirmed that H19 affects the poor prognosis in CRC. Knockdown of H19 dramatically reduces CRC cell proliferation through cell cycle control and migration. By microarray, as a non-biased approach and a bioinformatics technique, H19 affects RB protein and beta-catenin activity by regulating CDK8 expression (43).
HOX transcript antisense RNA (HOTAIR)
HOTAIR is 2,158 bp and is located on chromosome 12q13.13. HOTAIR was the first lncRNA to be reported as a transcriptional regulator and has also been reported to correlate with polycomb repressive complex 2 (PRC2) (44). Several reports have shown that HOTAIR has essential oncogenic function in breast cancer, pancreatic cancer, liver cancer, gastric cancer, and esophageal cancer (45,46). In CRC, the first report by Kogo et al. revealed that cancerous tissue has higher expression of HOTAIR than noncancerous tissue and that high HOTAIR expression correlates with liver metastasis. Furthermore, they showed that the reprogramming of PRC2 function influences HOTAIR expression (47). Following the first report, Svobada et al. evaluated the expression level of HOTAR in the blood of CRC patients, concluding that this lncRNA might be a prognostic marker in CRC (48). Depletion of HOTAIR is reported to increase E-cadherin expression and decrease matrix metalloproteinase 9, which indicates that HOTAIR is associated with the epithelial-mesenchymal transition (EMT). This study concluded that HOTAIR might promote CRC cell invasion and migration through the EMT (49). Yang et al. confirmed that HOTAIR influences the proliferation and invasion of CRC through a clinical analysis and in vitro experiments, and they found that HOTAIR also changes the radio-sensitivity of CRC (50). Bhan et al. reported that hypoxic conditions induce the expression of HOTAIR through the histone methylase MLL1 and HIF. In CRC, HIF1α, MLL1, and the histone acetylase p300 are abundant at the promotor site of HOTAIR, indicating that hypoxia-induced HOTAIR might correlate with the carcinogenesis of CRC (51). Thus, HOTAIR affects cancer phenotypes multi-directionally.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1)
MALAT-1, also known as nuclear enrichment autosomal transcript 2, is 8.7 kb and is located on chromosome 11q13.1. In several types of cancer, MALAT-1 is known to be associated with cancer metastasis and recurrence (52-55). The first MALAT-1 report that correlated it with CRC was reported by Zheng et al., which revealed that high expression of MALAT-1 is significantly associated with a poor prognosis in CRC and concluded that MALAT-1 might be a negative prognostic marker in CRC (56). Mechanistically, MALAT-1 is thought to influence the tumor microenvironment in CRC. Tumor-associated dendritic cells (TADCs), which secrete inflammatory cytokines, regulate migration, invasion, and the EMT. Cancer cell invasion and migration in the CRC cell lines SW480 and SW620 are increased through MALAT-1 overexpression with TADC-conditioned medium and chemokine (C-C motif) ligand 5 (57). Using microarray, Jie et al. found that MALAT-1 regulates miR-129-5p, inducing high expression of high motility group box protein 1 (HMGB1). Their results concluded that the MALAT-1/miR-129-5p/HMGB1 axis induces cancer development in CRC (58).
Nuclear-enriched abundant transcript 1 (NEAT1)
NEAT1 is a novel lncRNA that has two isoforms: NEAT1_1 and NEAT1_2. NEAT-1 was identified as a component of nuclear paraspeckles and was reported to regulate transcription (59,60). Li et al. evaluated NEAT-1 expression in CRC using CRC clinical samples and showed that high expression level of NEAT-1 was an independent prognostic factor of a poor outcome (61). Although there are few reports of NEAT-1 in CRC, the current study revealed a relationship between NEAT-1 and Akt signaling. Knockdown of NEAT-1 in CRC cell lines resulted in cell growth arrest and cell apoptosis by suppressing the phosphorylation of AKT at Ser473 (62).
Growth arrest-specific 5 (GAS5)
GAS5 regulates cell growth arrest by blocking target gene expression through activation of glucocorticoid receptor (63). GAS5 has been studied extensively, and recent reports have shown that this gene has important roles in cancer phenotypes by affecting proliferation, the EMT, and apoptosis (64,65). GAS5 has been identified as a biomarker in bladder cancer, non-small cell lung cancer, breast cancer, hepatocellular carcinoma, ovarian cancer, gastric cancer, cervical cancer, and head and neck squamous cell carcinoma (66-72). In CRC, Yin et al. revealed its clinical significance by evaluating CRC samples. Furthermore, they showed that overexpression of GAS5 inhibits cell proliferation in vitro and in vivo (73). Interestingly, another study by Kong et al. found that GAS5 and the lncRNA Yiya correlate with a poor prognosis and predict the risk of liver metastasis in early-stage CRC patients (74). Moreover, GAS5 was revealed to inhibit interleukin-10 and vascular endothelial growth factor expression, resulting in suppression of CRC carcinogenesis. Because few reports have described the relationship between lncRNA and angiogenesis, this study could provide insights into the development of CRC diagnosis and therapy (75). Thus, GAS5 broadly regulates CRC phenotypes and is a potential biomarker and therapeutic target.
LincRNA-p21
LincRNA-p21 is a P53 transcriptional target that represses the expression of its target genes by associating with hnRNP-K (12). Functionally, two reports have shown an association between LincRNA-p21 and β-catenin signaling (76,77). Wang et al. revealed that LincRNA-p21 increases radio-sensitivity, which is induced through the suppression of β-catenin (76). Another study showed that the tumorigenicity of CRC stem cells is mitigated by LincRNA-p21 through suppression of β-catenin signaling and concluded that LincRNA-p21 could be a possible target for cancer stem cell-targeting therapy (77).
Promoter of CDKN1A antisense DNA damage-activated RNA (PANDAR)
PANDAR is located at 6p21.2 and is upregulated by doxorubicin (78). Chen et al. determined the mechanisms of PANDAR in CRC. Changes in PANDAR expression could not alter proliferation, apoptosis, and senescence. However, using curcumin-treated CRC cells, knockdown of PANDAR induced apoptosis and increased senescence (79). Lu et al. reported that high expression of PANDAR is related to a poor prognosis in CRC and promotes metastasis via the EMT (80).
Taurine-upregulated gene 1 (TUG1)
TUG1 was initially reported to be associated with eye development in mice (81). Additionally, dysregulation of TUG1 regulates carcinogenesis in a variety of tumors (82-84). Regarding CRC, two studies showed that TUG1 promotes invasion and migration via the EMT pathway. These studies also concluded that knockdown of TUG1 could be a therapeutic strategy for CRC (85). Li et al. noted that TUG1 regulates the sensitivity of CRC cells to methotrexate (MTX). Knockdown of TUG1 re-sensitized the MTX resistance in MTX-resistant CRC cell lines. Furthermore, using a bioinformatics technique, they revealed that TUG1 binds directly to miRNA-186 and regulates its target, CPEB2 (86).
TINCR
TINCR is a 3.7-kb lncRNA and has been reported as a suppressor of CRC metastasis. Zuo-Yang revealed the clinical significance and mechanisms of TINCR in CRC. Evaluating cancer tissue and adjacent tissues in 44 CRC patients, they found significantly lower expression of TINCR in CRC samples. Additionally, they observed that the loss of TINCR induced Wnt-β-catenin signaling via hydrolysis of EpCAM (87). As with CCAT2, SNP status might influence the development of CRC. Yongbin et al. found that genetic variations in TINCR might contribute to the susceptibility to and progression of CRC, analyzing three SNPs in TINCR. They concluded that two SNPs, rs2288947 and rs8105637, might be independent biomarkers that are related to the occurrence and progression of CRC (88).
Concluding remarks and future perspectives
Surgical techniques and chemotherapies have improved the outcomes of CRC patients. Especially, anti-PD-L1 therapy has been reported to be effective in solid tumors, and the US Food and Drug Administration has approved the anti-PD-L1 agent nivolumab (OPDIVO, Bristol-Myers Squibb Company) for CRC patients with mismatch repair-deficient (dMMR) and microsatellite instability-high (MSI-H) metastatic CRC that has progressed following combination treatment with fluoropyrimidine, oxaliplatin, and irinotecan. Thus, new therapeutic strategies for CRC have appeared and had favorable results; however, they are thought to remain insufficient.
Clinical studies that are targeting miRNA are still going. For example, miravirsen, which is a nucleic acid-modified DNA phosphorothioate oligonucleotide complementary to miR-122, suppresses hepatitis C virus (HCV) RNA and clear phase I trials and is now in phase II trials (89). In cancer, miR-34 mimics (MRX34) against primary liver carcinoma, solid tumors, and hematological malignancies have been evaluated in a phase I trial, but this clinical study has been terminated, due to immune-related severe adverse events (ClinicalTrials.gov). Furthermore, since miRNAs are short RNAs (~22 nucleotides), one miRNA targets numerous genes, indicating that it is difficult for miRNA-based drugs to target specific oncogenes or tumor suppressors. Thus, new therapies targeting miRNAs remain underdeveloped. In contrast, lncRNAs bind to chromatin, DNA, RNA, and proteins directly or indirectly, which indicates that the target genes of lncRNAs might be specific compared with those of miRNAs (90).
Although carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9) are broadly used as CRC markers, more specific biomarkers are eagerly anticipated. As mentioned above, many studies have reported the possibility of using lncRNA as biomarkers for several types of cancers, including CRC, in tumor samples and blood samples. Like H19, some lncRNAs are abundant in and specific to CRC, rendering them good biomarkers for CRC. Intriguingly, Liang et al. found that lncRNA expression profiles discriminate several types of microbes in the gut (91). These results indicate that fecal lncRNAs could be possible biomarkers for screening CRC.
Other than the representative oncogenic lncRNAs described above, several lncRNAs contribute to proliferation, invasion, and metastasis in CRC, which could aid its early diagnosis. As possible targets of CRC, many studies have focused on determining the clinical importance of lncRNAs. Several clinical studies targeting lncRNAs have just begun. Hence, further research is necessary to identify the oncogenic mechanisms and pathways of lncRNAs. lncRNAs affect the hallmarks of CRC, implying that they will undoubtedly lead to discoveries that are relevant to the diagnosis and treatment of CRC.
Acknowledgements
We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29.[Crossref][PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7.[Crossref][PubMed]
- Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015;34:5003-11.[Crossref][PubMed]
- Higgs PG, Lehman N. The RNA World: molecular cooperation at the origins of life. Nat Rev Genet 2015;16:7-17.[Crossref][PubMed]
- Tragante V, Moore JH, Asselbergs FW. The ENCODE project and perspectives on pathways. Genet Epidemiol 2014;38:275-80.[Crossref][PubMed]
- Chodroff RA, Goodstadt L, Sirey TM, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol 2010;11:R72.[Crossref][PubMed]
- Ulitsky I, Shkumatava A, Jan CH, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147:1537-50.[Crossref][PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208.[Crossref][PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915-27.[Crossref][PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307.[Crossref][PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61.[Crossref][PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19.[Crossref][PubMed]
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120-4.[Crossref][PubMed]
- Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 2010;24:2264-9.[Crossref][PubMed]
- Sigova AA, Abraham BJ, Ji X, et al. Transcription factor trapping by RNA in gene regulatory elements. Science 2015;350:978-81.[Crossref][PubMed]
- Simon MD, Pinter SF, Fang R, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 2013;504:465-9.[Crossref][PubMed]
- Chu C, Zhang QC, da Rocha ST, et al. Systematic discovery of Xist RNA binding proteins. Cell 2015;161:404-16.[Crossref][PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38.[Crossref][PubMed]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011;470:284-8.[Crossref][PubMed]
- Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013;493:231-5.[Crossref][PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-463.[Crossref][PubMed]
- Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011;29:742-9.[Crossref][PubMed]
- Feng Y, Hu X, Zhang Y, et al. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol 2014;1165:115-43.[Crossref][PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74.[Crossref][PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905.[Crossref][PubMed]
- Kim T, Cui R, Jeon YJ, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A 2014;111:4173-8.[Crossref][PubMed]
- McCleland ML, Mesh K, Lorenzana E, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest 2016;126:639-52.[Crossref][PubMed]
- Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014;24:513-31.[Crossref][PubMed]
- Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61.[Crossref][PubMed]
- Redis RS, Vela LE, Lu W, et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell 2016;61:520-34.[Crossref][PubMed]
- Ozawa T, Matsuyama T, Toiyama Y, et al. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 'gene desert', serve as important prognostic biomarkers in colorectal cancer. Ann Oncol 2017;28:1882-8.[Crossref][PubMed]
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991;351:153-5.[Crossref][PubMed]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987;51:987-1000.[Crossref][PubMed]
- Lustig O, Ariel I, Ilan J, et al. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev 1994;38:239-46.[Crossref][PubMed]
- Tabano S, Colapietro P, Cetin I, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics 2010;5:313-24.[Crossref][PubMed]
- Byun HM, Wong HL, Birnstein EA, et al. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res 2007;67:10753-8.[Crossref][PubMed]
- Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005;280:29625-36.[Crossref][PubMed]
- Kim SJ, Park SE, Lee C, et al. Alterations in promoter usage and expression levels of insulin-like growth factor-II and H19 genes in cervical carcinoma exhibiting biallelic expression of IGF-II. Biochim Biophys Acta 2002;1586:307-15.[Crossref][PubMed]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007;13:313-6.[Crossref][PubMed]
- Tsang WP, Ng EK, Ng SS, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010;31:350-8.[Crossref][PubMed]
- Chen S, Bu D, Ma Y, et al. H19 Overexpression Induces Resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in Colon Cancer Cells. Neoplasia 2017;19:226-36.[Crossref][PubMed]
- Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 2016;7:22159-73.[PubMed]
- Ohtsuka M, Ling H, Ivan C, et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-beta-Catenin Signaling in Colorectal Cancer. EBioMedicine 2016;13:113-24.[Crossref][PubMed]
- Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013;32:1616-25.[Crossref][PubMed]
- Ishibashi M, Kogo R, Shibata K, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep 2013;29:946-50.[Crossref][PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6.[Crossref][PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6.[Crossref][PubMed]
- Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014;35:1510-5.[Crossref][PubMed]
- Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 2014;32:395-402.[Crossref][PubMed]
- Yang XD, Xu HT, Xu XH, et al. Knockdown of long non-coding RNA HOTAIR inhibits proliferation and invasiveness and improves radiosensitivity in colorectal cancer. Oncol Rep 2016;35:479-87.[Crossref][PubMed]
- Bhan A, Deb P, Shihabeddin N, et al. Histone methylase MLL1 coordinates with HIF and regulate lncRNA HOTAIR expression under hypoxia. Gene 2017;629:16-28.[Crossref][PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41.[Crossref][PubMed]
- Yamada K, Kano J, Tsunoda H, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci 2006;97:106-12.[Crossref][PubMed]
- Lin R, Maeda S, Liu C, et al. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007;26:851-8.[Crossref][PubMed]
- Tseng JJ, Hsieh YT, Hsu SL, et al. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod 2009;15:725-31.[Crossref][PubMed]
- Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol 2014;7:3174-81.[PubMed]
- Kan JY, Wu DC, Yu FJ, et al. Chemokine (C-C Motif) Ligand 5 is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression Through Non-Coding RNA MALAT-1. J Cell Physiol 2015;230:1883-94.[Crossref][PubMed]
- Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol 2017. [Epub ahead of print].[Crossref][PubMed]
- Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717-26.[Crossref][PubMed]
- Hirose T, Virnicchi G, Tanigawa A, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014;25:169-83.[Crossref][PubMed]
- Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015;6:27641-50.[PubMed]
- Peng W, Wang Z, Fan H. LncRNA NEAT1 Impacts Cell Proliferation and Apoptosis of Colorectal Cancer via Regulation of Akt Signaling. Pathol Oncol Res 2017;23:651-6.[Crossref][PubMed]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8.[Crossref][PubMed]
- Tan Q, Zuo J, Qiu S, et al. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding RNA, plasma, GAS5, biomarker. Int J Oncol 2017;50:1729-38.[Crossref][PubMed]
- Tao H, Zhang JG, Qin RH, et al. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology 2017;386:11-8.[Crossref][PubMed]
- Zhang H, Guo Y, Song Y, et al. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol 2017;79:49-55.[Crossref][PubMed]
- Wu Y, Lyu H, Liu H, et al. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep 2016;6:31093.[Crossref][PubMed]
- Chang L, Li C, Lan T, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep 2016;13:1541-50.[Crossref][PubMed]
- Gao J, Liu M, Zou Y, et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep 2015;34:3212-21.[Crossref][PubMed]
- Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014;14:319.[Crossref][PubMed]
- Cao S, Liu W, Li F, et al. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol 2014;7:6776-83.[PubMed]
- Gee HE, Buffa FM, Camps C, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer 2011;104:1168-77.[Crossref][PubMed]
- Yin D, He X, Zhang E, et al. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol 2014;31:253.[Crossref][PubMed]
- Kong H, Wu Y, Zhu M, et al. Long non-coding RNAs: novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget 2016;7:50428-36.[Crossref][PubMed]
- Li Y, Li Y, Huang S, et al. Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget 2017;8:13690-702.[PubMed]
- Wang G, Li Z, Zhao Q, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol Rep 2014;31:1839-45.[Crossref][PubMed]
- Wang J, Lei ZJ, Guo Y, et al. miRNA-regulated delivery of lincRNA-p21 suppresses beta-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget 2015;6:37852-70.[PubMed]
- Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621-9.[Crossref][PubMed]
- Chen T, Yang P, Wang H, et al. Silence of long noncoding RNA PANDAR switches low-dose curcumin-induced senescence to apoptosis in colorectal cancer cells. Onco Targets Ther 2017;10:483-91.[Crossref][PubMed]
- Lu M, Liu Z, Li B, et al. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol 2017;143:71-81.[Crossref][PubMed]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005;15:501-12.[Crossref][PubMed]
- Han Y, Liu Y, Gui Y, et al. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol 2013;107:555-9.[Crossref][PubMed]
- Zhang Q, Geng PL, Yin P, et al. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev 2013;14:2311-5.[Crossref][PubMed]
- Xu Y, Wang J, Qiu M, et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol 2015;36:1643-51.[Crossref][PubMed]
- Wang L, Zhao Z, Feng W, et al. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget 2016;7:51713-9.[PubMed]
- Li C, Gao Y, Li Y, et al. TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis. Biochem Biophys Res Commun 2017;491:552-7.[Crossref][PubMed]
- Zhang ZY, Lu YX, Zhang ZY, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget 2016;7:22639-49.[PubMed]
- Zheng Y, Yang C, Tong S, et al. Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget 2017;8:33536-43.[PubMed]
- Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013;368:1685-94.[Crossref][PubMed]
- Murphy MB, Medvedev AE. Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity. J Leukoc Biol 2016;99:839-50.[Crossref][PubMed]
- Liang L, Ai L, Qian J, et al. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci Rep 2015;5:11763.[Crossref][PubMed]
Cite this article as: Ohtsuka M, Tanemura M, Akamatsu H. Long noncoding RNAs regulate malignant phenotypes in colorectal cancer. Biotarget 2018;2:4.


