
Divergent functions of a ribosome maturation factor
The ribosome, the large macromolecular complex responsible for the translation of messenger RNA (mRNA) into proteins, must be matured and assembled correctly for effective function. Generally, ribosome maturation and assembly involves (I) translation of ribosomal proteins, and transcription and processing of ribosomal RNA (rRNA); (II) modification of ribosomal proteins and rRNA; (III) shaping and remodeling of rRNA in parallel with ribosomal protein recruitment (1). These processes are reliant on assembly factors that include nucleases, rRNA-modifying enzymes, GTPases, RNA helicases and chaperones (1,2). Dysregulation of this process has been implicated in a wide variety of human diseases.
In general, protein translation and ribosomal assembly is less well studied for mitochondrial ribosomes (mitoribosomes) than bacterial and eukaryotic ribosomes. Recent high-resolution electron cryomicroscopy (cryo-EM) structures have revealed considerable evolutionary divergences of mitoribosomes from their bacterial ancestors both in structure and composition (3-7). For example, the mammalian mitoribosome, which consists of the 39S large subunit (mt-LSU) and 28S small subunit (mt-SSU), has 36 additional mitoribosome specific proteins compared to the bacterial ribosome and considerable shortening of rRNA (6,8). However, the core scaffold of rRNA retains essential features such as the peptidyl transferase centre (PTC) and the messenger RNA decoding site. The assembly of human mitoribosomes involves the coordination of 82 nuclear-encoded mitochondrial proteins with mitochondrially encoded RNA (1 mt-tRNA and 2 mt-rRNA; 12S of the mt-SSU and 16S of the mt-LSU), and is dependent on nuclear-encoded assembly factors. Many mitoribosomal maturation and assembly factors have bacterial homologs, but given the increased complexity of mitoribosomal assembly, additional factors unique to the mitoribosome have been found (9-18).
In a recent Biochemical Journal article by Rozanska et al. entitled “The human RNA-binding protein RBFA promotes the maturation of the mitochondrial ribosome”, the authors examined the role that RBFA plays in the assembly of human mitoribosomes (19). RBFA is homologous to bacterial RbfA, and both contain an RNA-binding ‘KH’ domain. However, the puzzle is that while bacterial RbfA is involved in the processing of the 5’ region of 17S rRNA precursor to form mature 16S rRNA (20), human mt-rRNA is excised from polycistronic transcripts without the need for further processing. Thereby alternative roles are likely to have evolved for these structurally related proteins. Consistent with this, RBFA has acquired considerable N- and C-terminal extensions and is over twice the size of its bacterial counterpart.
To study the cellular role of RBFA the authors first demonstrated that RBFA associates preferentially with the mt-SSU. By sequencing RNA isolated by crosslinking immunoprecipitation (CLIP-seq) they found that RBFA associated largely with helices 44 and 45 at the 3’ end of 12S rRNA. Helix 44 is an integral part of the decoding centres and forms several inter-subunit bridges. Helix 45 harbours two highly conserved dimethylated adenines in a tetraloop that tucks into a groove of h44 close to the decoding centre (Figure 1). These are two of only ten nucleotides of mt-rRNA that have been found to be post-transcriptionally modified in mammals (21). This is modest compared to eukaryotic cytoplasmic and bacterial ribosomes which have in excess of 200 and 30 modifications, respectively (21). In bacteria, loss of helix 45 dimethylation has been shown to decrease translational fidelity (22). Knockout of the methyltransferase enzyme responsible for these modifications, TFB1M, is embryonically lethal in mice and heterozygosity showed impaired pancreatic islet cell mitochondrial function contributing to the pathogenesis of type II diabetes (23,24).
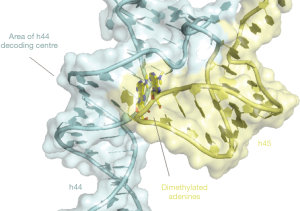
ERAL1, the GTPase and 12S mt-rRNA chaperone, also binds to this site (12), but is not functionally interchangeable with RBFA in human cell lines. The authors were therefore interested to see the effect of RBFA and ERAL1 on the methylation status of h45. To achieve this, they performed parallel immunoprecipitations of RBFA and ERAL1-FLAG, and determined the methylation status of the extracted rRNA. The results showed that RBFA was mostly bound to dimethylated h45 while ERAL1 was largely bound to unmethylated h45. It is likely that ERAL1 and RBFA bind 12S rRNA sequentially, as mass spectrometry data did not detect a significant proportion of ERAL1 co-immunoprecipitating with RBFA, and the CLIP-protected RNA fragments were found to be similar for the two proteins.
The next step was to look for newly synthesized 12S rRNA to ascertain the effect of RBFA on promoting modification and maturation. To do this the authors first decreased the level of mature modified mt-SSU by depletion of ERAL1. Loss of ERAL1 has been shown to lead to depletion of nascent 12S mt-rRNA (12). This was followed by repletion of ERAL1 with either concomitant RBFA depletion or a non-depleted control. The modification status of the rRNA was then evaluated. They found that when 12S rRNA was depleted together with RBFA the number of unmodified nucleotides was substantially increased compared to the non-depleted control, indicative of RBFA playing a role in promoting 12S rRNA methylation. The authors propose that RBFA functions by pushing h45 out to expose the adjacent adenines to its methyltransferase, TFB1M. This is consistent with a low-resolution cryo-EM structure (25) that shows RbfA displaces h44 and the adjacent area of h45 by ~25 Å when bound to the bacterial SSU (25). However, the exact molecular mechanism by which RBFA influences the modification process remains speculative.
The authors then demonstrated that unmodified 12S rRNA could be incorporated into mt-SSU, but only mt-SSU with the correctly modified and matured 12S rRNA forms monosomes. Thus, this paper shows that RBFA plays a role in promoting dimethylation of the 12S rRNA of the mt-SSU, and this is an important quality control step in monosome formation. Binding of RBFA to h44/45 may promote dimethylation of highly conserved consecutive adenines by increasing accessibility of these residues to their methyltransferase TFB1M. It does not appear that the dimethylation is a prerequisite for assembly of the mt-SSU. Instead the dimethylation appears instrumental in mt-rRNA maturation and functional monosome formation.
In summary, the authors have shown an example of a known ribosomal maturation factor acquiring alternative functions to support ribosome maturation as an adaptive strategy, most likely resultant of the evolutionary divergence of the mitoribosome.
Acknowledgments
Thanks to A Brown and JL Llácer for their help.
Funding: N Desai is funded by the Wellcome Trust Clinical PhD Fellowship (110301/Z/15/Z).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Editor-in-Chief Dr. Maorong Jiang (Laboratory Animal Center, Nantong University, Nantong, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2017.11.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem 2011;80:501-26. [Crossref] [PubMed]
- De Silva D, Tu YT, Amunts A, et al. Mitochondrial ribosome assembly in health and disease. Cell Cycle 2015;14:2226-50. [Crossref] [PubMed]
- Desai N, Brown A, Amunts A, et al. The structure of the yeast mitochondrial ribosome. Science 2017;355:528-31. [Crossref] [PubMed]
- Brown A, Amunts A, Bai XC, et al. Structure of the large ribosomal subunit from human mitochondria. Science 2014;346:718-22. [Crossref] [PubMed]
- Greber BJ, Boehringer D, Leibundgut M, et al. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 2014;515:283-6. [PubMed]
- Amunts A, Brown A, Toots J, et al. Ribosome. The structure of the human mitochondrial ribosome. Science 2015;348:95-8. [Crossref] [PubMed]
- Amunts A, Brown A, Bai XC, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014;343:1485-9. [Crossref] [PubMed]
- Greber BJ, Bieri P, Leibundgut M, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 2015;348:303-8. [Crossref] [PubMed]
- Metodiev MD, Spåhr H, Loguercio Polosa P, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 2014;10:e1004110 [Crossref] [PubMed]
- Spåhr H, Habermann B, Gustafsson CM, et al. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci U S A 2012;109:15253-8. [Crossref] [PubMed]
- Kolanczyk M, Pech M, Zemojtel T, et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol Biol Cell 2011;22:1-11. [Crossref] [PubMed]
- Dennerlein S, Rozanska A, Wydro M, et al. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J 2010;430:551-8. [Crossref] [PubMed]
- Rorbach J, Boesch P, Gammage PA, et al. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell 2014;25:2542-55. [Crossref] [PubMed]
- Dalla Rosa I, Durigon R, Pearce SF, et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res 2014;42:8500-15. [Crossref] [PubMed]
- Tu YT, Barrientos A. The Human Mitochondrial DEAD-Box Protein DDX28 Resides in RNA Granules and Functions in Mitoribosome Assembly. Cell Rep 2015; [Epub ahead of print]. [Crossref] [PubMed]
- He J, Cooper HM, Reyes A, et al. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res 2012;40:6097-108. [Crossref] [PubMed]
- Brown A, Rathore S, Kimanius D, et al. Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol 2017;24:866-9. [Crossref] [PubMed]
- Antonicka H, Shoubridge EA. Mitochondrial RNA Granules Are Centers for Posttranscriptional RNA Processing and Ribosome Biogenesis. Cell Rep 2015; [Epub ahead of print]. [Crossref] [PubMed]
- Rozanska A, Richter-Dennerlein R, Rorbach J, et al. The human RNA-binding protein RBFA promotes the maturation of the mitochondrial ribosome. Biochem J 2017;474:2145-58. [Crossref] [PubMed]
- Bylund GO, Wipemo LC, Lundberg LA, et al. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol 1998;180:73-82. [PubMed]
- Bohnsack MT, Sloan KE. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell Mol Life Sci 2017; [Epub ahead of print]. [Crossref] [PubMed]
- van Buul CP, Visser W, van Knippenberg PH. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett 1984;177:119-24. [Crossref] [PubMed]
- Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet 2003;33:23-4. [Crossref] [PubMed]
- Koeck T, Olsson AH, Nitert MD, et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab 2011;13:80-91. [Crossref] [PubMed]
- Datta PP, Wilson DN, Kawazoe M, et al. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell 2007;28:434-45. [Crossref] [PubMed]
Cite this article as: Desai N. Divergent functions of a ribosome maturation factor. Biotarget 2017;1:17.


