
Mitochondrial ribosome-binding factor A (mtRBFA): mediator of rRNA methylation and maturation
Mitochondria are organelles perhaps most famous for energy production by oxidative phosphorylation (OXPHOS). In addition to producing ATP, mitochondria are multi-faceted organelles that mediate processes such as autophagy, differentiation, calcium homeostasis, and the immune response (1,2). Mitochondrial dysfunction has been implicated in a number of diseases, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and diabetes, to name a few (3). Understanding mitochondrial function is critical to finding cures for these diseases. Although the field of mitochondrial biology has progressed significantly in the past decade, a number of questions remain unanswered, specifically regarding the logistics of mammalian mitochondrial gene expression.
Mitochondria contain a circular genome, and mitochondrial gene expression refers to transcription, RNA processing, and translation of mitochondrial DNA. The mitochondrial genome is much smaller than the nuclear genome, containing about 16,000 base pairs of DNA in mammals. Mitochondrial DNA encodes 13 genes required for OXPHOS, as well as two rRNAs and 22 tRNAs. Although the majority of proteins required for mitochondrial function are imported into the organelle, the 13 OXPHOS subunits encoded within the mitochondrial genome (7 subunits of complex I, 1 subunit from complex III, 3 subunits of complex IV, and 2 subunits of ATP synthase) are critical for the function of their respective complexes (4). Few or no nucleotides separate genes within mitochondrial DNA, and the lack of non-coding regions negates the need for extensive post-transcriptional processing.
Mitochondria-encoded OXPHOS genes are transcribed by mitochondrial transcription machinery and translated into proteins by dedicated mitochondrial ribosomes (4). All proteins that participate in mitochondrial transcription and translation are encoded in the nucleus and imported into the organelle. In addition to nuclear-encoded protein subunits, the eukaryotic mitochondrial ribosome contains rRNA that is encoded in mitochondrial DNA. This requires close coordination of protein import into mitochondria and transcription of mitochondrial rRNA. The fully assembled mitochondrial ribosome has a sedimentation coefficient of 55S and is composed of a large 39S subunit and a small 28S subunit (5). The 55S ribosome contains 80 nuclear-encoded proteins and three ribosomal RNA molecules: 16S large subunit rRNA, 12S small subunit rRNA, and a large subunit tRNA (mt-tRNAVal) (6). In the recent paper by Rozanska et al. published in Biochemical Journal, ribosome-binding factor A (RBFA) has been identified as a protein that binds to the 12S rRNA and promotes its assembly into the mature ribosome (7).
Mitochondrial ribosome assembly describes the process by which mitochondrial rRNA is transcribed and processed and associates with imported ribosomal proteins to form large and small ribosomal subunits. The large and small subunits then join together to form a functional 55S monosome (8). This process occurs in mitochondria within the matrix. Although ribosome assembly for eukaryotic cytosolic ribosomes requires >170 accessory proteins, this process in bacteria and mitochondria is much less complex (8,9).
The mitochondrial genome is composed of a heavy strand and a light strand, named such due to their buoyancy in cesium chloride gradients (10). The heavy strand encodes both rRNAs, as well as 14 tRNAs and 12 OXPHOS subunits. Mitochondrial transcription requires a minimum of three transcription factors: mtRNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM), and mitochondrial transcription factor B2 (TFB2M) (11). Transcription of the heavy strand by POLRMT initiates from one of two promoter regions. The first promoter (HSP1) initiates upstream of tRNAPhe and terminates after 12S and 16S rRNAs. The second promoter (HSP2) initiates upstream of the 12S rRNA and its transcription results in a long polycistronic molecule containing all of the mRNAs and most of the tRNAs encoded on the heavy strand (12). Processing of mitochondrial RNA requires excision of the tRNAs, leaving leaderless mRNAs and rRNAs. mRNAs are polyadenylated, but rRNAs have the addition of only 1 or 2 adenines (13).
Mitochondrial rRNA undergoes a number of post-transcriptional modifications required for assembly into the functional ribosome. These modifications include base methylation, 2'-O-ribose methylation and the incorporation of the modified nucleotide pseudouridine (13). The 12S rRNA contains 5 base methylations, including 2 adenines (A936/A937 near the 3' end) that are conserved in all domains of life (14). Methylation of rRNA at A936/A937 is performed by mitochondrial transcription factor B1 (TFB1M), a methyltransferase that is a homolog of the bacterial methyltransferase KsgA (14-16).
In addition to rRNA modification proteins, E. coli requires an essential ribosome assembly factor, RBFA. This protein is critical for growth adaptation to cold temperatures and interacts with the 5' terminal helix region of the small rRNA (8,17). RBFA assists in the processing of the 17S rRNA precursor into the 16S mature rRNA. Since mitochondrial rRNAs do not require extensive processing, eukaryotic mitochondrial RBFA has until this point been functionally undefined. Although the NMR structure of mitochondrial RBFA has been solved (PDB 2E7G), it was labeled as a “putative ribosome-binding factor”. Work by Rozanska et al. has now elucidated a role for mitochondrial RBFA in ribosome assembly (7).
Another essential assembly factor in E. coli is the Era protein, a GTPase that binds near the 3' end of the 16S rRNA and prevents base pairing of the Shine-Dalgarno sequence with the rRNA. Era also induces conformational changes in the 16S rRNA that may increase activity of the methyltransferase KsgA (18). Mitochondrial ERAL1 is a homologue of the E. coli Era protein. ERAL1 knockdown reduces mitochondrial protein synthesis and leads to reduced stability of 12S rRNA (19).
In the manuscript by Rozanska et al., the authors first performed a sequence and structural alignment of mitochondrial RBFA with E. coli RBFA. Although the two proteins share structural similarity, sequence similarity was lacking. Subcellular fractionation studies revealed that RBFA in HEK293 cells was indeed localized to mitochondria. Sucrose density gradient centrifugation was used to determine whether RBFA associates with individual ribosomal subunits or with the intact 55S monosome. In this assay, researchers found RBFA free at the top of the gradient and near the middle of the gradient bound to the small ribosomal subunit. RBFA was not present in later fractions with the 39S large subunit or the 55S intact ribosome, although mS29 and mS40 were not present in the 55S fractions either. To confirm whether RBFA binds to large or small subunits, immunoprecipitation of small ribosomal protein mS27 pulled down RBFA in gradient fractions with small subunit proteins but not in fractions with large subunits or intact ribosomes. Taken together, this data indicates that RBFA is not associated with mature 55S ribosomes and must leave prior to subunit joining.
Within the small subunit, the authors confirmed the exact nucleotides of 12S rRNA bound by RBFA using a complex immunoprecipitation assay called CLIP (20). This method, based on UV crosslinking of proteins bound to RNA, allows sequencing of small RNA pieces covered by the protein. Results from this experiment indicated that RBFA binds to mitochondrial 12S rRNA near the 3' end. This is in contrast to E. coli RBFA, which binds at the 5' end of bacterial rRNA. The nucleotides covered by RBFA binding are near the neck region of the small subunit, shown in Figure 1, and overlap with the region of rRNA bound by ERAL1. Included in the RBFA-covered nucleotides were the highly conserved dimethylated adenines A936/A937 (shown in orange in Figure 1).
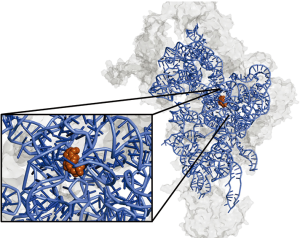
To determine the importance of RBFA for cellular growth, Rozanska et al. knocked it down using siRNA. This impeded growth of both HeLa and HEK293 cell lines. The authors next set out to determine whether reductions in levels of mtDNA (leading to reductions in 12S rRNA) would affect gene expression of RBFA. In mtDNA-depleted Rho0 cells, RBFA could not be detected and the cells continued to grow normally. This suggests that cells lacking mtDNA might be less dependent on RBFA. In fact, Rho0 cells seemed to grow better in the absence of RBFA, a phenomenon that warrants further investigation.
Although overexpression of Era in E. coli can partially rescue RBFA knockdown, expression of ERAL1 in HEK293 cells did not rescue the RBFA-induced growth defect. In the reciprocal experiment, RBFA overexpression was unable to rescue reduced 12S rRNA levels observed with ERAL1 siRNA. RBFA knockdown also did not affect mitochondrial protein synthesis, as measured by western blot against mitochondrial OXPHOS subunits and an 35S mitochondrial translation assay. Since RBFA knockdown had such a drastic effect on growth, it is surprising that the growth failure was not due to defects in mitochondrial translation.
The final (and arguably the most critical) experiment performed to explore the function of RBFA was a primer read-through assay to measure rRNA methylation. Since part of the RBFA coverage region of the 12S rRNA included 2 conserved adenine nucleotides that must be methylated in mature 12S rRNA, perhaps RBFA contributes to this process. In the primer read-through assay, modification of the rRNA at A936/A937 caused termination of reverse transcription and resulted in a band in the middle of the toe print. If the modification was not present, the enzyme “read-through” those nucleotides and stopped at a later position, leading to a larger oligonucleotide and a band at the top of the toe print. In the absence of RBFA, less methylation (more read-through) was observed, suggesting a role for this protein in stimulating methylation. In conjunction with this experiment, the authors also quantified the amount of methylation in RNA that immunoprecipitated with ERAL1 and RBFA. ERAL1-associated rRNA was methylated less than that associated with RBFA, indicating that ERAL1 binds to RNA before RBFA.
Finally, the authors looked at the amount of modified rRNA bound to the small subunit by immunoprecipitation. Although siRNA knockdown of RBFA led to reduced methylation of 12S rRNA within the 28S small subunit, it did not affect methylation of 12S rRNA bound within the 55S ribosome. Thus, some sort of discrimination must be taking place (independent of RBFA) to prevent unmodified rRNA from being incorporated into the intact 55S ribosome. It would be interesting to see whether loss of RBFA and subsequent reduction of A936/A937 dimethylation affects binding of translation initiation factor 3 (IF3) to the small subunit, since IF3 binds to this region of helix 45 in bacteria.
To determine how RBFA might be affecting methylation of rRNA, the authors performed a western blot for ERAL1 and TFB1M. Both ERAL1 and TFB1M were increased in the absence of RBFA. Since methylation of A936/A937 is reduced in RBFA knockdown cells, it is surprising to also see increased levels of TFB1M (the methyltransferase credited for performing those modifications) in these cells. The authors attribute this phenomenon to stimulation of alternative mechanisms to methylate A936/A937. They postulate that RBFA may bind behind helices 44 and 45 and push A936/A937 outward to expose them for methylation. If this is the case, increased TFB1M may reflect a cellular feedback loop in response to reduced accessibility and methylation of A936/A937 and the potential for insufficient ribosome supply. Additional work is needed to elucidate the exact mechanism of methylation stimulated by RBFA, but the involvement of TFB1M should not be excluded.
This work has obvious importance in identifying a new protein that contributes to ribosome assembly in mitochondria. However, since mitochondrial RBFA (mtRBFA) appears to be dispensable for mitochondrial translation, further work is needed to connect growth defects to reduced levels of RBFA. Since E. coli RBFA participates in the cold-shock response, it would be interesting to see whether the mtRBFA pathway becomes more critical to translation during temperature stress in mammalian cells.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Editor-in-Chief Dr. Maorong Jiang (Laboratory Animal Center, Nantong University, Nantong, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2017.11.02). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity 2015;42:406-17. [Crossref] [PubMed]
- Brookes PS, Yoon Y, Robotham JL, et al. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol 2004;287:C817-33. [Crossref] [PubMed]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012;148:1145-59. [Crossref] [PubMed]
- Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 2012;1819:1035-54.
- O'Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life 2003;55:505-13. [Crossref] [PubMed]
- Amunts A, Brown A, Toots J, et al. Ribosome. The structure of the human mitochondrial ribosome. Science 2015;348:95-8. [Crossref] [PubMed]
- Rozanska A, Richter-Dennerlein R, Rorbach J, et al. The human RNA-binding protein RBFA promotes the maturation of the mitochondrial ribosome. Biochem J 2017;474:2145-58. [Crossref] [PubMed]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem 2011;80:501-26. [Crossref] [PubMed]
- Fromont-Racine M, Senger B, Saveanu C, et al. Ribosome assembly in eukaryotes. Gene 2003;313:17-42. [Crossref] [PubMed]
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1999;1410:103-23. [Crossref] [PubMed]
- Arnold JJ, Smidansky ED, Moustafa IM, et al. Human mitochondrial RNA polymerase: structure-function, mechanism and inhibition. Biochim Biophys Acta 2012;1819:948-60.
- Lodeiro MF, Uchida A, Bestwick M, et al. Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc Natl Acad Sci U S A 2012;109:6513-8. [Crossref] [PubMed]
- Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem J 2012;444:357-73. [Crossref] [PubMed]
- De Silva D, Tu YT, Amunts A, et al. Mitochondrial ribosome assembly in health and disease. Cell Cycle 2015;14:2226-50. [Crossref] [PubMed]
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011;2:611-31. [Crossref] [PubMed]
- Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res 2007;35:4042-54. [Crossref] [PubMed]
- Uchiumi T, Ohgaki K, Yagi M, et al. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res 2010;38:5554-68. [Crossref] [PubMed]
- Tu C, Zhou X, Tarasov SG, et al. The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA. Proc Natl Acad Sci U S A 2011;108:10156-61. [Crossref] [PubMed]
- Dennerlein S, Rozanska A, Wydro M, et al. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J 2010;430:551-8. [Crossref] [PubMed]
- Darnell R. CLIP (cross-linking and immunoprecipitation) identification of RNAs bound by a specific protein. Cold Spring Harb Protoc 2012;2012:1146-60.
Cite this article as: Christian BE. Mitochondrial ribosome-binding factor A (mtRBFA): mediator of rRNA methylation and maturation. Biotarget 2017;1:16.


