
Vacuolar H+-ATPases (V-ATPases) as therapeutic targets: a brief review and recent developments
Vacuolar H+-ATPases (V-ATPases)
V-ATPases are rotary motors that utilize hydrolysis of ATP to pump protons across a membrane against an electrochemical gradient (1). They are essential housekeeping enzymes in eukaryotic cells, where they acidify compartments of the endocytic and exocytic pathways, including late endosomes, lysosomes and compartments for uncoupling receptor and ligands. V-ATPases in the Golgi are vital for glycosylation and membrane trafficking (2). In addition to the housekeeping functions, some cells have layered upon the essential V-ATPases, additional V-ATPases that carry on specialized functions. These include acidifying an extracellular resorption compartment to solubilize bone mineral during bone resorption by osteoclasts (3), providing a driving force for loading neurotransmitters in neurons (4), and pumping protons across the plasma membrane of renal intercalated and proximal tubule cells to help maintain systemic pH homeostasis (5). Cancer cells also have plasma membrane V-ATPases that support their ability to survive and metastasize (6).
Major advances in understanding the structure, function and regulation of V-ATPases have been made during the past decade. Much of the core structure is known at the atomic level through high-resolution electron microscopy and crystal structures (7). Thirteen different subunits make up the “core” eukaryotic enzyme (Figure 1). These include eight subunits of the peripheral V1 complex and five subunits of the integral V0 complex. The stoichiometry of the V1 is 3A:3B:1C:1D:3E:3G:1F:1H. The stoichiometry of V0 is 1a:1b:5c:1d:1f. In addition to the isoforms listed, there are splice variants of some of the subunits (8). With the various isoforms and splice variants, the diversity of V-ATPases is high, although many questions remain regarding how these variations influence the ability of V-ATPases to carry out their various roles in cells.
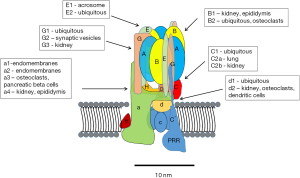
In addition to the core subunits, two accessory proteins have been identified AC45 (ATP6AP1) (9,10) and the (pro) renin receptor (PRR; also known as ATP6AP2) (11-14). AC45 regulates of V-ATPases in certain tissues. The PRR was first identified associated with the V-ATPase isolated from bovine chromaffin granules and called M8-9 (15), which was subsequently found to be from a gene in a chromosomal area linked to X-linked cone-rod dystrophy (16). The PRR gene, however, was not responsible for the pathology. Later this gene was identified as the “long sought” PRR, a protein that binds and activates (pro) renin, and enhances the activity of renin (17).
There are now strong evidences that the PRR is an essential chaperone required for the assembly and targeting of V-ATPase subunits (14,18-20). It is also an essential component of the canonical and non-canonical planar cell polarity WNT pathways (11,21,22). The PRR interacts with the c and d subunits of V-ATPases (23). Despite its name and activity, there is controversy regarding whether the ability to activate (pro) renin is of physiological importance (14).
V-ATPase-binding proteins and integration with cellular physiology
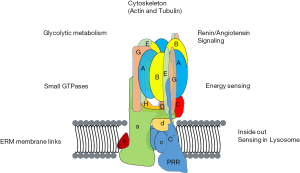
Numerous proteins bind to one or more V-ATPase subunit and this represents a means by which V-ATPases integrate into overall cellular physiology. The PRR interacts with a large number of proteins, including flamingo, cadherin EGF LAG seven-pass G-type receptor, frizzled, low-density lipoprotein receptor-related protein 6, (pro)renin, wingless-related integration site, phosphatase of regenerating liver-1, actin-binding LIM protein 2, CapZ alpha 2, pyruvate dehydrogenase E1, partitioning defective 3, and dihydrolipoamide acetyltransferase (E2) (11,24-27). In addition to PRR-binding proteins, a number of other proteins interact with V-ATPases. These include actin-filaments (28-31), aldolase and other glycolytic enzymes (32,33), NHE-RF and other PDZ-domain containing proteins (34), and ARNO (cytohesin 2) and Arf6 (35,36). Inside out amino acid sensing involving V-ATPases in association with mTorc1 and the Ragulator complex was demonstrated several years ago (37,38). Very recently, a study supporting a role for V-ATPase in energy sensing appeared. Binding of Lamtor1 and the Ragulator complex was shown to be regulated by the presence of active aldolase bound to V-ATPase (39). V-ATPases are also the target of proteins that are elements of infectious agents. The papillomavirus E5 oncoprotein interacts with V-ATPase, which is involved in cellular transformation in cervical cancer (40).
In general, data support the conception of V-ATPases as proton pumps that integrate with various processes in cells through multiple routes (Figure 2). The PDZ-binding domain found in the B1-isoform of B-subunit, but not B2, suggests a way in which specialized V-ATPases are controlled (34). In most cases, the mechanism of specialized usage of V-ATPases is unclear. The integration of V-ATPases into overall cell physiology implies that disruption of V-ATPases must be carefully managed because consequences beyond disruption of acidification of luminal compartments (which is itself profoundly disruptive) can be expected.
Pioneering efforts to selectively-inhibit V-ATPases
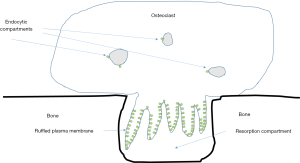
Bafilomycins, macrolide antibiotics were the first relatively specific inhibitors of V-ATPases identified (41). Soon thereafter, V-ATPase activity was shown to be required for bone resorption by osteoclasts (3). Osteoclasts express a much-studied example of specialized V-ATPases. In addition to the low levels of housekeeping enzymes found in all nucleated eukaryotic cells, osteoclasts also express a specialized subset of V-ATPases, present at 10–100 time higher levels than the housekeeping enzymes, which are transported to a subdomain domain of the plasma membrane called the ruffled plasma membrane or ruffled border (Figure 3) (42). The ruffled border targeted V-ATPases appear identical to housekeeping V-ATPases except that they contain isoforms of the a-subunit and d-subunit that are only expressed in osteoclasts and a few other few cell types.
The vital role of V-ATPases in bone resorption immediately suggested that a V-ATPase inhibitor might be an ideal agent to treat osteoporosis and other bone diseases that involve excess bone resorption. V-ATPases are also important for cancer cell metastasis, identifying cancer as another important pathology in which V-ATPases represent possible therapeutic targets (6,43). Unfortunately, bafilomycins are toxic to animals. This was expected given the central role of V-ATPases in acidification of vesicles and organelles. As mentioned above, two elements of the V-ATPases involved in bone resorption differ from the housekeeping enzymes; the a3-isoform of a-subunit and the d2-isoform of d-subunit, which have distributions that are more restricted (44-46). Thus, specific inhibitors for osteoclast V-ATPases might be possible.
Incentive to produce osteoclast specific inhibitors soon increased. Mutations in the a3- and d2- subunits triggered a form of osteopetrosis in which resorption was reduced (d2) (47) or blocked (a3) (48), and osteoblast bone formation occurred at a higher rate than normal. In patients with a3-mutated, this resulted in catastrophic uncoupling of bone remodeling leading to the disease autosomal malignant osteopetrosis (49,50). However, a therapeutic agent or strategy that blocked the activity of a3- or d2-containing V-ATPases might be selective enough to avoid systemic injuries in adults while triggering bone anabolic effects (51,52). Such an approach might treat osteoporosis better than the current standard of care.
Osteoclast V-ATPase selective inhibitors have been reported. The first was a bafilomycin derivative called SB 242784 (53). Testing of SB 242784 in the rat ovariectomized model for post-menopausal osteoporosis suggested that it was effective blocking bone loss, but that it required high doses to be effective. No overt adverse effects were reported over a 6-month period. Importantly, careful testing failed to detect disruption in urinary function (53). Despite this positive start, SB 242784 has not advanced further in pre-clinical or clinical studies.
FR167356, a V-ATPase inhibitor with a novel chemical structure, was reported to selectively inhibit osteoclast V-ATPases (54). It also blocked bone loss in the ovariectomized rat model, and at very high doses exhibited some of the expected anabolic effects. As with SB 242784, no disruption of kidney function was detected, even though FR167356 did not demonstrate the selectivity for the osteoclast V-ATPases compared with kidney V-ATPases that was displayed by SB 242784. Some concerns arise because rodent kidneys may not reflect damage that would occur to the human kidney (55).
Additional non-selective inhibitors have been reported. These include small molecules diphyllins (56), archazolids (57), salicylihalamide A (58), cleistanthin-A (59), iejimalides (60), and tulipaline A (61) and the protein inhibitor, Pea Albumin 1 subunit b (62). A number of studies have reported that bafilomycin and other non-selective V-ATPase inhibitors may be useful for treatment of certain types of cancers (63-70). To date these have not advanced into the clinic. Usefulness of non-selective V-ATPase inhibitors in cancer will likely require localized delivery and maintenance of the inhibitor at the site of the tumor. There is some evidence that certain types of cancer, like cancers with low cathepsin D activity, may be especially vulnerable to inhibition of V-ATPases (71). In such cases, a therapeutic window may exist for non-selective V-ATPase inhibitors.
There is strong evidence that certain types of cancers express plasma membrane V-ATPases and that these V-ATPases contain atypical V-ATPase subunits like the a3- and a4-isoforms of a-subunit (72). Targeting these isoforms or special characteristics associated with cancer V-ATPases might be therapeutically useful.
Rational approaches to generating selective inhibitors of V-ATPases
Selective inhibitors of V-ATPases in bone and certain cancers could prove of great clinical benefit. However, with the lack of success of efforts to develop selective inhibitors of V-ATPases as therapeutic agents despite extensive efforts, it is not clear that such agents will ever emerge from traditional screens. Increasing knowledge of the overall structure and mechanism of action evokes pessimism that selective inhibitors of the proton pumping activity in osteoclasts or cancer cells are possible. Rational approaches to selectively inhibiting V-ATPases based on structural features or secondary activities known to be found in specific subtypes of V-ATPases related to diseases, have recently been explored.
Disruption of subunit-subunit interactions between the V-ATPase isoforms found selectively in osteoclasts.
A strategy, taken by Morris Manolson’s lab at the University of Toronto, involved identifying inhibitors of protein-binding interactions between the V-ATPase subunits selectively expressed in osteoclast V-ATPases, a3 and d2, without perturbing interactions between other isoforms of the d-subunit and a-subunit (73). To accomplish this, in vitro protein-protein interactions between the a-subunit and d-subunit isoforms with each other or other V-ATPase subunit-binding partners were identified by yeast two-hybrid assays (74). Solid phase medium throughput assays were then developed for screening small molecule inhibitors of the interactions that had been identified. From these studies, inhibitors of binding between the a3 and B2 subunits and a3 and d2 subunits were identified, that had less effects on interaction between the other isoforms of a-subunit and d-subunit, and thus would be predicted to be selective for osteoclasts (Figure 4).
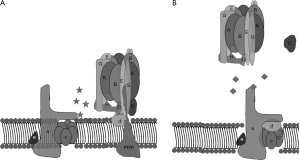
An inhibitor of the a3-B2 interaction, 3,4-dihydroxy-N’-(2-hydroxybenzylidene)benzohydrazide, was characterized in cell culture (74). As expected, this molecule strongly inhibited bone resorption, at concentrations where it did not significantly reduce the viability of osteoclasts, or the differentiation of precursors into osteoclasts. However, in vivo testing of this molecule has not yet been reported.
In similar screens, the molecule luteolin was identified as a selective inhibitor of binding between a3 and d2 (75). Luteolin is a flavone found in leaves, rinds, barks, clover blossoms and ragweed pollen. It is in dietary staples including celery, broccoli, green pepper, parsley, dandelion, chamomile tea, carrots, olive oil, peppermint, rosemary, navel oranges, oregano, and other sources (76). Unlike 3,4-dihydroxy-N’-(2-hydroxybenzylidene)benzohydrazide, luteolin had been previously proposed to have therapeutic benefits and several mechanisms advanced prior to the its identification as an inhibitor of binding between a3 and d2 (76). Proposed mechanisms of action included inhibition of cAMP and cGMP phosphodiesterase activity, as an anti-oxidant, by inhibition of NF kappa B-based pathways (the precise mechanism is unclear), by inhibition of cyclooxygenase 2, and by triggering apoptosis through generation of reactive oxygen species. Luteolin was shown, prior to work on V-ATPase, to inhibit osteoclasts, reduce bone loss in rats after ovariectomy and prevent titanium-induced bone loss (77).
Manolson’s group demonstrated inhibition of bone resorption without inhibition of cell survival, osteoclast formation or effects on V-ATPase gene expression (75). They suggested that this effect was due to disrupting the integrity of those V-ATPases that contain a3 and d2, which they propose are specifically those involved in the specialized function of bone resorption. Their results contrasted with previous studies, which reported inhibition of osteoclast formation due to disruption of receptor activator of nuclear factor kappa B-ligand signaling, which involves the NF kappa B pathway (77). As with luteolin, the ability of the osteoclasts from the d2-knockout osteoclasts to resorb bone was reduced (47). Luteolin is currently being investigated for use in the treatment of ailments including psoriatic arthritis, asthma, multiple sclerosis, and cancer, but there have been no recent developments in the bone field (78).
Disruption of binding between the B2-subunit of V-ATPase and microfilaments
Beth Lee, Stephen Gluck and Shannon Holliday reported that V-ATPases were isolated bound to microfilaments in osteoclasts, but not in various other cells and tissues (29). They proposed that because the binding was restricted to osteoclasts, it might be part of the mechanism that leads to the formation of the ruffled plasma membrane, the unusual V-ATPase-rich plasma membrane domain required for osteoclast activity. The Holliday lab went on to locate and characterize the microfilament-binding site in the B2-subunit and provide evidence that this site is required for V-ATPase transport to the ruffled membrane in cell culture experiments (28,79,80).
With that basis, a virtual screen seeking small molecules predicted to bind the microfilament-binding surface on the B2-subunit and competitively inhibit its binding to microfilaments was performed (Figure 5) (81). This approach involved making an atomic homology model of the V-ATPase subunit B in Swiss-Model protein structure homology modeling using the SWISS-MODEL workspace, based on the human sequence gi|13938355. Approximately 140,000 small molecules in a National Cancer Center Developmental Therapeutics Program repository were positioned in pockets of the actin-binding site in B2 using DOCKv6.1.0. Small molecules identified as having a high probability of binding the actin-binding site in B2 were then tested for their ability to block interaction between recombinant B2-subunit and microfilaments in binding assays. Among the small molecules identified was enoxacin, a second-generation fluoroquinolone antibiotic, which had been removed from the market in the United States (82,83). This small molecule could be obtained in bulk, had been tested extensively in humans, and remains FDA approved for use in humans. It continues to be used as an antibiotic in many countries, and has a reasonable, though far from spotless, safety profile (83). Like other fluoroquinolones, it has been associated with tendon tears and other side effects.
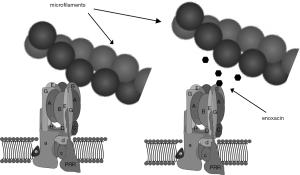
In vitro studies showed that enoxacin blocked both osteoclast differentiation and bone resorption by mature osteoclasts (81). These inhibitions did not involve triggering apoptosis (84). Although differentiation of multinuclear cells expressing tartrate resistant acid phosphatase (TRAP) activity was blocked in cell culture, expression of a variety of osteoclast selective proteins, like the a3-subunit of V-ATPase, was not affected (84). In the case of TRAP 5b, the enzyme responsible for the high levels of TRAP activity characteristic of osteoclasts, the mRNA and protein were expressed at normal levels, but activation of the proform of TRAP 5b by proteolytic cleavage did not occur. This result and other data suggested that enoxacin disrupts specialized vesicular trafficking necessary for osteoclast maturation and bone resorption, a result consistent with previous studies examining the role of the V-ATPase-microfilament binding interaction (85).
Enoxacin inhibited titanium-induced osteolysis in a preclinical model (86). However, bone loss was not significantly–inhibited in ovariectomized rat model of osteoporosis (L. S. Holliday, T. J. Wronski, E. J. Toro and D. A. Ostrov, unpub.). The half-life of enoxacin is relatively short in vivo and bone penetration is modest (87). Conjugation of enoxacin and other fluoroquinolones to bisphosphonates had been described as a part of an effort to make them more useful for the treatment of osteolysis due to bone infections (88). Bisphosphonates and their conjugates quickly accumulate at sites of active bone remodeling (89). In principle, the bisphosphonate-ester of enoxacin (bis-enoxacin), when introduced into animals should quickly accumulate to block osteoclast activity. Based on this reasoning, bis-enoxacin was synthesized by the Holliday lab in collaboration with SynQuest Laboratories (Alachua, FL, USA) (90).
Bis-enoxacin, like enoxacin, blocked binding between the B2-subunit and microfilaments in in vitro assays, and blocked osteoclast differentiation and bone resorption (90). It was more effective than enoxacin as an inhibitor of bone resorption in vitro, presumably because of the increased local concentration associated with the bone surface, which could be mobilized as osteoclasts began to resorb the bone.
Bis-enoxacin has been tested in three pre-clinical models. It was shown to block orthodontic tooth movement, which requires mechanically induced bone resorption, in a rat model (90). Bis-enoxacin blocked alveolar bone resorption associated with periodontitis in a rat model. In addition to blocking periodontitis-associated bone loss as efficiently as alendronate (91), bis-enoxacin and alendronate significantly reduced systemic oxidative stress associated with periodontal infections (92). Most importantly, bis-enoxacin was recently compared with zoledronate (a standard of care bisphosphonate used for osteoporosis) in a rat model of osteoporosis (93). Both molecules reduced bone loss. However, the bone composition of ovariectomized rats treated with bis-enoxacin was more like sham-ovariectomized animals than bones from the zoledronate treated animals. The proteoglycan content was higher bis-enoxacin-treated animals. Vitally, the strength of the bones and their resistance to fracture was better in the animals treated with bis-enoxacin compared with zoledronate. This shows that the composition of the bone is altered by bis-enoxacin in a beneficial way (93).
Based on these data, bis-enoxacin, or therapeutics with the same mechanism, represent a different class of potential anti-osteoporosis therapeutics, which reduce loss of bone mineral density and stimulate the formation of bone, which is intrinsically stronger and more resistant to fractures. Currently standard of care molecules (like zoledronate and the anti-RANKL inhibitor Denosumab) have reduced osteoporotic fracture rates significantly. However, there are still millions of fractures each year in the United States, and the reduction in fracture risk attributable to the current therapeutics has flattened out in recent years (94,95). A different type of therapeutic agent (like bis-enoxacin?) will be required to further reduce or eliminate osteoporotic fractures. It is possible that bis-enoxacin will work synergistically with other resorption inhibitors, but that has not yet been tested.
Enoxacin, bis-enoxacin, cancer and amyotrophic lateral sclerosis (ALS)
Concurrent with reports that enoxacin blocks binding between V-ATPase and microfilaments, other groups independently identified enoxacin as a stimulator of RNA interference and microRNA activity (96,97). The finding that microRNA activity was stimulated suggested a novel approach to treating cancer. MicroRNAs are a class of small RNAs that bind to specific mRNAs and block their expression (98). A general reduction in microRNA activity, which allows the expression of oncogenic proteins, was proposed to be important in oncogenesis (99). It follows that general stimulation, as reported to be achieved by enoxacin, might disrupt cancer cells and tumor formation. A collaboration involving the labs of Manel Estellar, George Calin, Carlo Croce and Raghu Kalluri, reported that enoxacin blocked the growth of colorectal cells in vitro and tumor metastasis and progression in a xenografted mouse model (100). They reported increases in microRNA activity were associated with interaction between enoxacin and TAR RNA binding protein. A subsequent study by Stamenkovic and colleagues showed that enoxacin is useful in the treatment of Ewing Sarcoma in a mouse model (101). They also found that it works synergistically with doxorubicin. The use of enoxacin to treat cancer in humans has not yet been reported.
Following similar logic, the Hornstein group reported that enoxacin (which crosses the blood brain barrier) is beneficial for neuromuscular function in two mouse models of ALS (102). They had found that down regulation of microRNA expression is a common denominator of multiple forms of human ALS.
Taken together with the bone studies, there is increasing evidence that enoxacin, bis-enoxacin or molecules with similar activity, may be useful to use in the treatment of important human diseases. However, several competing mechanisms to explain the therapeutic action have been advanced. Further progress may require an understanding of the activity that produces the therapeutic effects.
Mechanisms of enoxacin/bis-enoxacin
Multiple mechanisms have been proposed to explain the therapeutic effects of enoxacin. As described above, enoxacin first emerged from screens seeking inhibitors of binding between V-ATPase and microfilaments and stimulators of microRNAs. In addition, enoxacin suppresses the JNK signaling pathway (86), and induces alternative splicing of MdmX transcript and rescue of p53 activity (103). The RNA helicase DHX9 and PIWIL3 have also recently been identified as possible therapeutic targets of enoxacin (104). Another possibility is that, enoxacin and bis-enoxacin, by binding the V-ATPase B2-subunit, may disrupt interactions with the stator arms composed of E and G subunits. If this required higher concentrations of enoxacin, it could explain high concentration effects of enoxacin in killing cancer (and other) cells.
It is possible that most or all of these activities are required for the therapeutic effects of enoxacin. In this case, enoxacin or very similar molecules may be uniquely capable of having the therapeutic effects that have been documented. Strategies would then depend on delivering or targeting the molecule to specific locations, as with bis-enoxacin targeting enoxacin to bone. Another possibility is that all of the detected activities have a common origin. For example, it is known that V-ATPases are integrated into various metabolic and physiologic pathways. Perhaps disruption of targeting of V-ATPases by enoxacin leads to alterations in microRNA levels and other signaling pathways. Alternatively, it is plausible that stimulation of microRNAs, rescue of p53 or stimulation of the JNK pathway could have a variety of cellular effects. We think that it is informative that enoxacin and bis-enoxacin inhibit osteoclast formation and bone resorption in cell culture at concentrations where we do not detect significant increases in microRNA activity in osteoclasts (Zuo and Holliday, unpublished observation). In addition, we identified another inhibitor of the V-ATPase-microfilament binding interaction, directed toward another part of the actin-binding pocket on the B2-subunit, with a completely different structure compared with enoxacin that exhibited similar or identical effects on osteoclast formation and bone resorption (81). It is unlikely that both molecules also directly stimulate microRNAs. Nevertheless, additional research is required to establish the mechanism of enoxacin’s therapeutic effects with respect to different pathologies. We believe at this point that the crucial conclusion is that enoxacin and bis-enoxacin work by different mechanisms than the current therapeutic agents that are used to treat bone diseases, cancer and ALS, and may have benefits either alone or in combination other therapeutics. Synergistic effects between enoxacin and doxorubicin demonstrated for the treatment of Ewing Sarcoma in a preclinical model support this idea (101).
Virtual similarity based screening identifies new V-ATPase inhibitors
Rational approaches to identifying V-ATPase inhibitors have yielded agents like luteolin and enoxacin that have intriguing therapeutic characteristics, but multiple mechanisms of action. To identify novel agents with similar activity or analogs to these agents with higher therapeutic activity, which of the known mechanistic activities should be used to screen. For example, a more potent inhibitor of microfilament-B-subunit interaction might be identified, but if that activity is not important for the therapeutic effect, it would have no therapeutic value. An approach to this problem is to perform virtual screens seeking molecules with similar structures to known therapeutics with desired properties. This is a computational, lead molecule approach that can utilize multiple lead molecules with different structures. Candidates can be narrowed from a very large library to a few molecules, which can then be screened directly against a target cell (or perhaps even a disease in some preclinical models). This approach is less concerned about underlying mechanism of action and instead focuses on the desired therapeutic effect.
Results of such an approach to identifying V-ATPase-directed anti-cancer molecules recently appeared. Virtual screening was performed, directed by the structures of four disparate V-ATPase-inhibitors, enoxacin, diphyllin, benzimidazole analog and NIK-12192 (105). The virtual screen was performed assaying chemical compounds in the University of Cincinnati, Drug Discovery Center (UC DDC) Library, consisting of 362,000 compounds, and the National Cancer Institute (NCI) Small Molecule Repository (SMR), with 263,365 compounds. After identifying 429 compounds, manual screening of these by Lipinski’s rules and visual perception of relatedness to compounds of interest, selected 28 compounds. These were screened for activity against MDA-MB-231 and MDA-MB-468 cells. An initial lead molecule was identified and variations were synthesized. This process identified a novel bisbenzimidazole pharmacophore that has a mid-nanomolar IC50 toward MDA-MB-468 cells and represents the first V-ATPase inhibitor of its class (105).
Perspectives
Because of its central role in cellular physiology and pathophysiology, V-ATPase is an exciting target for therapeutic development, but one that is extremely challenging. Knockouts of isoforms of V-ATPases that are selectively expressed in osteoclasts and involved in bone remodeling lead to inhibition of bone resorption and increased bone formation (52). Unusual V-ATPases are vital for cancer growth and metastasis (6). Agents that block the V-ATPases involved in these pathologies are plausible routes to treating these diseases. Already, rationally identified molecules targeting V-ATPases, enoxacin and its bisphosphonate ester, bis-enoxacin, have been reported to have unique and beneficial activities for the treatment of bone disease and cancer in preclinical models (86,90,91,93,100,101). Increasing knowledge of V-ATPases, and how their structure, binding interactions, regulation in different physiologic circumstances, and the diversity of V-ATPases derived from subunit isoforms and splice variants, comes into play in generating their cell specific activities, should open new paths for the rational identification of V-ATPase-directed therapeutics. The potential of such agents warrants the efforts required to overcome the challenges incumbent in targeting a housekeeping enzyme for drug development.
Acknowledgments
The author would like to thank the many colleagues that have helped at various stages of his work on V-ATPases. He would especially like to acknowledge Stephen L. Gluck, University of California, San Francisco who introduced him to the enzyme. Beth S. Lee, Ohio State University, Ming Lu, University of California San Francisco, David A. Ostrov, University of Florida College of Medicine, Jian Zuo, University of Florida College of Dentistry, Shih-Hua Chen and Edgardo J. Toro played essential roles in his studies of V-ATPases and his lab’s efforts to develop V-ATPase-targeted drugs.
Funding: Dr. Holliday’s work on V-ATPases was funded by NIH-NIAMS R01 AR47959 (LSH) and NIH-NIDCR R21 DE19862 01A1 (LSH).
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/biotarget.2017.12.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cotter K, Stransky L, McGuire C, et al. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem.Sci 2015;40:611-22. [Crossref] [PubMed]
- Kornak U, Reynders E, Dimopoulou A, et al. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat Genet 2008;40:32-4. [Crossref] [PubMed]
- Blair HC, Teitelbaum SL, Ghiselli R, et al. Osteoclastic bone resorption by a polarized vacuolar proton pump. Science 1989;245:855-7. [Crossref] [PubMed]
- Moriyama Y, Maeda M, Futai M. The role of V-ATPase in neuronal and endocrine systems. J Exp Biol 1992;172:171-8. [PubMed]
- Gluck SL, Underhill DM, Iyori M, et al. Physiology and biochemistry of the kidney vacuolar H+-ATPase. Annu Rev Physiol 1996;58:427-45. [Crossref] [PubMed]
- Sennoune SR, Martinez-Zaguilan R, Vacuolar H. ATPase signaling pathway in cancer. Curr Protein Pept Sci 2012;13:152-63. [Crossref] [PubMed]
- Rawson S, Harrison MA, Muench SP. Rotating with the brakes on and other unresolved features of the vacuolar ATPase. Biochem Soc Trans 2016;44:851-5. [Crossref] [PubMed]
- Miranda KC, Karet FE, Brown D. An extended nomenclature for mammalian V-ATPase subunit genes and splice variants. PLoS One 2010;5:e9531 [Crossref] [PubMed]
- Feng H, Cheng T, Pavlos NJ, et al. Cytoplasmic terminus of vacuolar type proton pump accessory subunit Ac45 is required for proper interaction with V(0) domain subunits and efficient osteoclastic bone resorption. J Biol Chem 2008;283:13194-204. [Crossref] [PubMed]
- Jansen EJ, Scheenen WJ, Hafmans TG, et al. Accessory subunit Ac45 controls the V-ATPase in the regulated secretory pathway. Biochim Biophys Acta 2008;1783:2301-10.
- Cruciat CM, Ohkawara B, Acebron SP, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 2010;327:459-63. [Crossref] [PubMed]
- Kinouchi K, Ichihara A, Sano M, et al. The role of individual domains and the significance of shedding of ATP6AP2/(pro)renin receptor in vacuolar H(+)-ATPase biogenesis. PLoS ONE 2013;8:e78603 [Crossref] [PubMed]
- Müller DN, Binger KJ, Riediger F. Prorenin receptor regulates more than the renin-angiotensin system. Ann Med 2012;44:S43-8. [Crossref] [PubMed]
- Peters J. The (pro)renin receptor and its interaction partners. Pflugers Arch 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Ludwig J, Kerscher S, Brandt U, et al. Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J Biol Chem 1998;273:10939-47. [Crossref] [PubMed]
- Demirci FY, White NJ, Rigatti BW, et al. Identification, genomic structure, and screening of the vacuolar proton-ATPase membrane sector-associated protein M8-9 gene within the COD1 critical region (Xp11.4). Mol Vis 2001;7:234-9. [PubMed]
- Nguyen G, Delarue F, Burckle C, et al. Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J Clin Invest 2002;109:1417-27. [Crossref] [PubMed]
- Jansen EJ, Martens GJ. Novel insights into V-ATPase functioning: distinct roles for its accessory subunits ATP6AP1/Ac45 and ATP6AP2/(pro) renin receptor. Curr Protein Pept Sci 2012;13:124-33. [Crossref] [PubMed]
- Kinouchi K, Ichihara A, Sano M, et al. The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ Res 2010;107:30-4. [Crossref] [PubMed]
- Krop M, Lu X, Danser AH, et al. The (pro)renin receptor. A decade of research: what have we learned? Pflugers Arch 2013;465:87-97. [Crossref] [PubMed]
- Buechling T, Bartscherer K, Ohkawara B, et al. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr Biol 2010;20:1263-8. [Crossref] [PubMed]
- Sihn G, Rousselle A, Vilianovitch L, et al. Physiology of the (pro)renin receptor: Wnt of change? Kidney Int 2010;78:246-56. [Crossref] [PubMed]
- Kinouchi K, Ichihara A, Itoh H. Functional characterization of (pro)renin receptor in association with V-ATPase. Front Biosci (Landmark.Ed) 2011;16:3216-23.
- Hermle T, Petzoldt AG, Simons M. The role of proton transporters in epithelial Wnt signaling pathways. Pediatr Nephrol 2011;26:1523-7. [Crossref] [PubMed]
- Hermle T, Saltukoglu D, Grunewald J, et al. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr Biol 2010;20:1269-76. [Crossref] [PubMed]
- Funke-Kaiser H, Zollmann FS, Schefe JH, et al. Signal transduction of the (pro)renin receptor as a novel therapeutic target for preventing end-organ damage. Hypertens Res 2010;33:98-104. [Crossref] [PubMed]
- Kanda A. Atp6ap2/(Pro) renin Receptor is Required for Laminar Formation during Retinal Development in Mice. Nippon Ganka Gakkai Zasshi 2015;119:787-98. [PubMed]
- Holliday LS, Lu M, Lee BS, et al. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem 2000;275:32331-7. [Crossref] [PubMed]
- Lee BS, Gluck SL, Holliday LS. Interaction between vacuolar H(+)-ATPase and microfilaments during osteoclast activation. J Biol Chem 1999;274:29164-71. [Crossref] [PubMed]
- Vitavska O, Wieczorek H, Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem 2003;278:18499-505. [Crossref] [PubMed]
- Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem 2005;280:1070-6. [Crossref] [PubMed]
- Lu M, Holliday LS, Zhang L, et al. Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J Biol Chem 2001;276:30407-13. [Crossref] [PubMed]
- Lu M, Sautin YY, Holliday LS, et al. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem 2004;279:8732-9. [Crossref] [PubMed]
- Breton S, Wiederhold T, Marshansky V, et al. The B1 subunit of the H+ATPase is a PDZ domain-binding protein. Colocalization with NHE-RF in renal B-intercalated cells. J Biol Chem 2000;275:18219-24. [Crossref] [PubMed]
- Hosokawa H, Dip PV, Merkulova M, et al. The N termini of a-subunit isoforms are involved in signaling between vacuolar H+-ATPase (V-ATPase) and cytohesin-2. J Biol Chem 2013;288:5896-913. [Crossref] [PubMed]
- Hurtado-Lorenzo A, Skinner M, Annan JE, et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol 2006;8:124-36. [Crossref] [PubMed]
- Bar-Peled L, Schweitzer LD, Zoncu R, et al. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012;150:1196-208. [Crossref] [PubMed]
- Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678-83. [Crossref] [PubMed]
- Zhang CS, Hawley SA, Zong Y, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 2017;548:112-6. [Crossref] [PubMed]
- Skinner MA, Wildeman AG. beta(1) integrin binds the 16-kDa subunit of vacuolar H(+)-ATPase at a site important for human papillomavirus E5 and platelet-derived growth factor signaling. J Biol Chem 1999;274:23119-27. [Crossref] [PubMed]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A 1988;85:7972-6. [Crossref] [PubMed]
- Qin A, Cheng TS, Pavlos NJ, et al. V-ATPases in osteoclasts: Structure, function and potential inhibitors of bone resorption. Int J Biochem Cell Biol 2012;44:1422-35. [Crossref] [PubMed]
- Stransky L, Cotter K, Forgac M. The Function of V-ATPases in Cancer. Physiol Rev 2016;96:1071-91. [Crossref] [PubMed]
- Mattsson JP, Li X, Peng SB, et al. Properties of three isoforms of the 116-kDa subunit of vacuolar H+-ATPase from a single vertebrate species. Cloning, gene expression and protein characterization of functionally distinct isoforms in Gallus gallus. Eur J Biochem 2000;267:4115-26. [Crossref] [PubMed]
- Toyomura T, Oka T, Yamaguchi C, et al. Three subunit a isoforms of mouse vacuolar H(+)-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J Biol Chem 2000;275:8760-5. [Crossref] [PubMed]
- Smith AN, Jouret F, Bord S, et al. Vacuolar H+-ATPase d2 subunit: molecular characterization, developmental regulation, and localization to specialized proton pumps in kidney and bone. J Am Soc Nephrol 2005;16:1245-56. [Crossref] [PubMed]
- Lee SH, Rho J, Jeong D, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med 2006;12:1403-9. [Crossref] [PubMed]
- Li YP, Chen W, Liang Y, et al. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet 1999;23:447-51. [Crossref] [PubMed]
- Kornak U, Schulz A, Friedrich W, et al. Mutations in the a3 subunit of the vacuolar H(+)-ATPase cause infantile malignant osteopetrosis. Hum Mol Genet 2000;9:2059-63. [Crossref] [PubMed]
- Scimeca JC, Quincey D, Parrinello H, et al. Novel mutations in the TCIRG1 gene encoding the a3 subunit of the vacuolar proton pump in patients affected by infantile malignant osteopetrosis. Hum Mutat 2003;21:151-7. [Crossref] [PubMed]
- Henriksen K, Flores C, Thomsen JS, et al. Dissociation of Bone Resorption and Bone Formation in Adult Mice with a Non-Functional V-ATPase in Osteoclasts Leads to Increased Bone Strength. PLoS ONE 2011;6:e27482 [Crossref] [PubMed]
- Karsdal MA, Martin TJ, Bollerslev J, et al. Are nonresorbing osteoclasts sources of bone anabolic activity? J Bone Miner Res 2007;22:487-94. [Crossref] [PubMed]
- Price PA, June HH, Buckley JR, et al. SB 242784, a selective inhibitor of the osteoclastic V-H+ATPase, inhibits arterial calcification in the rat. Circ Res 2002;91:547-52. [Crossref] [PubMed]
- Niikura K, Takeshita N, Takano M. A vacuolar ATPase inhibitor, FR167356, prevents bone resorption in ovariectomized rats with high potency and specificity: potential for clinical application. J Bone Miner Res 2005;20:1579-88. [Crossref] [PubMed]
- Finberg KE, Wagner CA, Bailey MA, et al. The B1-subunit of the H(+) ATPase is required for maximal urinary acidification. Proc Natl Acad Sci U S A 2005;102:13616-21. [Crossref] [PubMed]
- Sørensen MG, Henriksen K, Neutzsky-Wulff AV, et al. Diphyllin, a Novel and Natural Potent V-ATPase Inhibitor, Abrogates Acidification of the Osteoclastic Resorption Lacunae and Bone Resorption. J Bone Miner Res 2007;22:1640-8. [Crossref] [PubMed]
- Huss M, Sasse F, Kunze B, et al. Archazolid and apicularen: novel specific V-ATPase inhibitors. BMC Biochem 2005;6:13. [Crossref] [PubMed]
- Wu Y, Liao X, Wang R, et al. Total synthesis and initial structure-function analysis of the potent V-ATPase inhibitors salicylihalamide A and related compounds. J Am Chem Soc 2002;124:3245-53. [Crossref] [PubMed]
- Zhao Y, Lu Y, Ma J, et al. Synthesis and Evaluation of Cleistanthin A Derivatives as Potent Vacuolar H(+) -ATPase Inhibitors. Chem Biol Drug Des 2015;86:691-6. [Crossref] [PubMed]
- Kazami S, Muroi M, Kawatani M, et al. Iejimalides show anti-osteoclast activity via V-ATPase inhibition. Biosci Biotechnol Biochem 2006;70:1364-70. [Crossref] [PubMed]
- Caboni P, Tronci L, Liori B, et al. Tulipaline A: structure-activity aspects as a nematicide and V-ATPase inhibitor. Pestic Biochem Physiol 2014;112:33-9. [Crossref] [PubMed]
- Chouabe C, Eyraud V, Da SP, et al. New mode of action for a knottin protein bioinsecticide: pea albumin 1 subunit b (PA1b) is the first peptidic inhibitor of V-ATPase. J Biol Chem 2011;286:36291-6. [Crossref] [PubMed]
- Lim JH, Park JW, Kim MS, et al. Bafilomycin induces the p21-mediated growth inhibition of cancer cells under hypoxic conditions by expressing hypoxia-inducible factor-1alpha. Mol Pharmacol 2006;70:1856-65. [Crossref] [PubMed]
- Avnet S, Di PG, Lemma S, et al. V-ATPase is a candidate therapeutic target for Ewing sarcoma. Biochim Biophys Acta 2013;1832:1105-16.
- De Milito A, Iessi E, Logozzi M, et al. Proton pump inhibitors induce apoptosis of human B-cell tumors through a caspase-independent mechanism involving reactive oxygen species. Cancer Res 2007;67:5408-17. [Crossref] [PubMed]
- Morimura T, Fujita K, Akita M, et al. The proton pump inhibitor inhibits cell growth and induces apoptosis in human hepatoblastoma. Pediatr Surg Int 2008;24:1087-94. [Crossref] [PubMed]
- Niikura K. Effect of a V-ATPase inhibitor, FR202126, in syngeneic mouse model of experimental bone metastasis. Cancer Chemother Pharmacol 2007;60:555-62. [Crossref] [PubMed]
- Sasazawa Y, Futamura Y, Tashiro E, et al. Vacuolar H+-ATPase inhibitors overcome Bcl-xL-mediated chemoresistance through restoration of a caspase-independent apoptotic pathway. Cancer Sci 2009;100:1460-7. [Crossref] [PubMed]
- Sennoune SR, Bakunts K, Martinez GM, et al. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol 2004;286:C1443-52. [Crossref] [PubMed]
- Shen W, Zou X, Chen M, et al. Effects of diphyllin as a novel V-ATPase inhibitor on gastric adenocarcinoma. Eur J Pharmacol 2011;667:330-8. [Crossref] [PubMed]
- Kitazawa S, Nishizawa S, Nakagawa H, et al. Cancer with low cathepsin D levels is susceptible to vacuolar (H+)-ATPase inhibition. Cancer Sci 2017;108:1185-93. [Crossref] [PubMed]
- McGuire C, Cotter K, Stransky L, et al. Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness. Biochim Biophys Acta 2016;1857:1213-8.
- Kartner N, Manolson MF. V-ATPase Subunit Interactions: The Long Road to Therapeutic Targeting. Curr Protein Pept Sci 2012;13:164-79. [Crossref] [PubMed]
- Kartner N, Yao Y, Li K, et al. Inhibition of osteoclast bone resorption by disrupting vacuolar H+-ATPase a3-B2 subunit interaction. J Biol Chem 2010;285:37476-90. [Crossref] [PubMed]
- Crasto GJ, Kartner N, Yao Y, et al. Luteolin inhibition of V-ATPase a3-d2 interaction decreases osteoclast resorptive activity. J Cell Biochem 2013;114:929-41. [Crossref] [PubMed]
- Luo Y, Shang P, Li D. Luteolin: A Flavonoid that Has Multiple Cardio-Protective Effects and Its Molecular Mechanisms. Front Pharmacol 2017;8:692. [Crossref] [PubMed]
- Shin DK, Kim MH, Lee SH, et al. Inhibitory effects of luteolin on titanium particle-induced osteolysis in a mouse model. Acta Biomater 2012;8:3524-31. [Crossref] [PubMed]
- Liu-Smith F, Meyskens FL. Molecular mechanisms of flavonoids in melanin synthesis and the potential for the prevention and treatment of melanoma. Mol Nutr Food Res 2016;60:1264-74. [Crossref] [PubMed]
- Chen SH, Bubb MR, Yarmola EG, et al. Vacuolar H+-ATPase binding to microfilaments: regulation in response to phosphatidylinositol 3-kinase activity and detailed characterization of the actin-binding site in subunit B. J Biol Chem 2004;279:7988-98. [Crossref] [PubMed]
- Zuo J, Jiang J, Chen SH, et al. Actin Binding Activity of Subunit B of Vacuolar H(+)-ATPase Is Involved in Its Targeting to Ruffled Membranes of Osteoclasts. J Bone Miner Res 2006;21:714-21. [Crossref] [PubMed]
- Ostrov DA, Magis AT, Wronski TJ, et al. Identification of enoxacin as an inhibitor of osteoclast formation and bone resorption by structure-based virtual screening. J Med Chem 2009;52:5144-51. [Crossref] [PubMed]
- Henwood JM, Monk JP. Enoxacin. A review of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs 1988;36:32-66. [Crossref] [PubMed]
- Rubinstein E. History of quinolones and their side effects. Chemotherapy 2001;47:3-8. [Crossref] [PubMed]
- Toro EJ, Zuo J, Ostrov DA, et al. Enoxacin directly inhibits osteoclastogenesis without inducing apoptosis. J Biol Chem 2012;287:17894-904. [Crossref] [PubMed]
- Toro EJ, Ostrov DA, Wronski TJ, et al. Rational Identification of Enoxacin as a Novel V-ATPase-Directed Osteoclast Inhibitor. Curr Protein Pept Sci 2012;13:180-91. [Crossref] [PubMed]
- Liu X, Qu X, Wu C, et al. The effect of enoxacin on osteoclastogenesis and reduction of titanium particle-induced osteolysis via suppression of JNK signaling pathway. Biomaterials 2014;35:5721-30. [Crossref] [PubMed]
- Fong IW, Rittenhouse BR, Simbul M, et al. Bone penetration of enoxacin in patients with and without osteomyelitis. Antimicrob Agents Chemother 1988;32:834-7. [Crossref] [PubMed]
- Herczegh P, Buxton TB, McPherson JC III, et al. Osteoadsorptive bisphosphonate derivatives of fluoroquinolone antibacterials. J Med Chem 2002;45:2338-41. [Crossref] [PubMed]
- Rogers MJ, Frith JC, Luckman SP, et al. Molecular mechanisms of action of bisphosphonates. Bone 1999;24:73S-9S. [Crossref] [PubMed]
- Toro EJ, Zuo J, Guiterrez A, et al. Bis-enoxacin Inhibits Bone Resorption and Orthodontic Tooth Movement. J Dent Res 2013;92:925-31. [Crossref] [PubMed]
- Rivera MF, Chukkapalli SS, Velsko IM, et al. Bis-enoxacin blocks rat alveolar bone resorption from experimental periodontitis. PLoS ONE 2014;9:e92119 [Crossref] [PubMed]
- Oktay S, Chukkapalli SS, Rivera-Kweh MF, et al. Periodontitis in rats induces systemic oxidative stress that is controlled by bone-targeted antiresorptives. J Periodontol 2015;86:137-45. [Crossref] [PubMed]
- Liu X, Qu X, Nie T, et al. The Beneficial Effects of Bisphosphonate-enoxacin on Cortical Bone Mass and Strength in Ovariectomized Rats. Front Pharmacol 2017;8:355. [Crossref] [PubMed]
- Black DM, Rosen CJ. Postmenopausal Osteoporosis. N Engl J Med 2016;374:2096-7. [Crossref] [PubMed]
- Fisher A, Martin J, Srikusalanukul W, et al. Bisphosphonate use and hip fracture epidemiology: ecologic proof from the contrary. Clin Interv Aging 2010;5:355-62. [Crossref] [PubMed]
- Zhang Q, Zhang C, Xi Z. Enhancement of RNAi by a small molecule antibiotic enoxacin. Cell Res 2008;18:1077-9. [Crossref] [PubMed]
- Shan G, Li Y, Zhang J, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat.Biotechnol 2008;26:933-40. [Crossref] [PubMed]
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol 2002;18:495-513. [Crossref] [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [Crossref] [PubMed]
- Melo S, Villanueva A, Moutinho C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A 2011;108:4394-9. [Crossref] [PubMed]
- Cornaz-Buros S, Riggi N, DeVito C, et al. Targeting cancer stem-like cells as an approach to defeating cellular heterogeneity in Ewing sarcoma. Cancer Res 2014;74:6610-22. [Crossref] [PubMed]
- Emde A, Eitan C, Liou LL, et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J 2015;34:2633-51. [Crossref] [PubMed]
- Valianatos G, Valcikova B, Growkova K, et al. A small molecule drug promoting miRNA processing induces alternative splicing of MdmX transcript and rescues p53 activity in human cancer cells overexpressing MdmX protein. PLoS ONE 2017;12:e0185801 [Crossref] [PubMed]
- Cao S, Sun R, Wang W, et al. RNA helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am J Transl Res 2017;9:674-82. [PubMed]
- Patil R, Kulshrestha A, Tikoo A, et al. Identification of Novel Bisbenzimidazole Derivatives as Anticancer Vacuolar (H(+))-ATPase Inhibitors. Molecules 2017;22:E1559 [Crossref] [PubMed]
Cite this article as: Holliday LS. Vacuolar H+-ATPases (V-ATPases) as therapeutic targets: a brief review and recent developments. Biotarget 2017;1:18.


